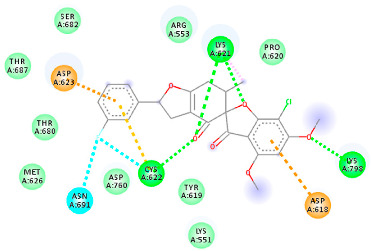
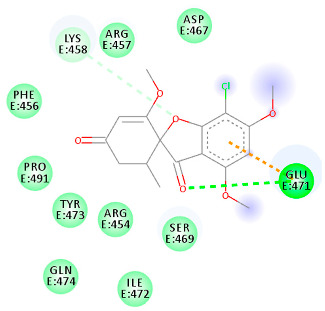
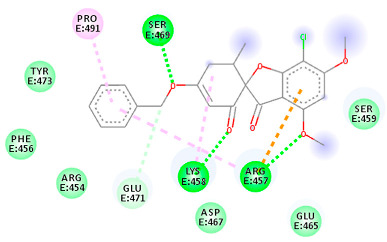
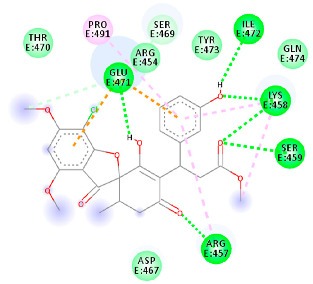
Table 1.
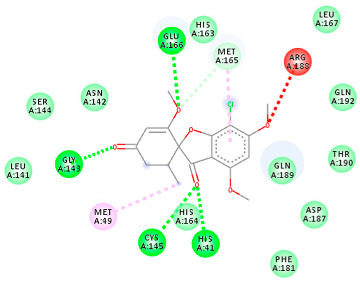
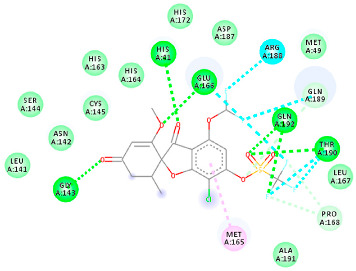
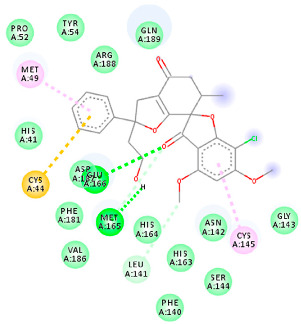
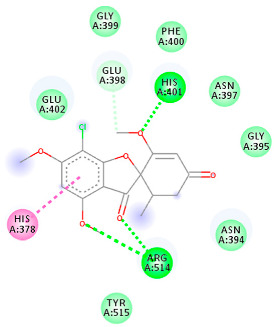
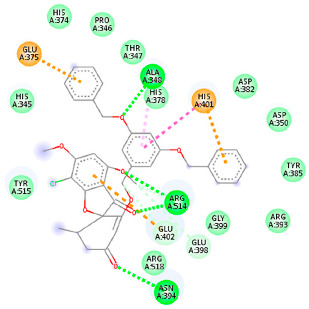
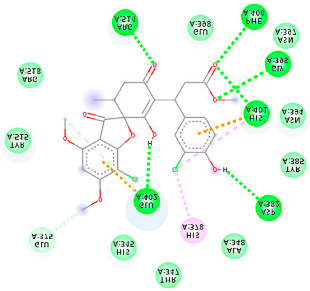
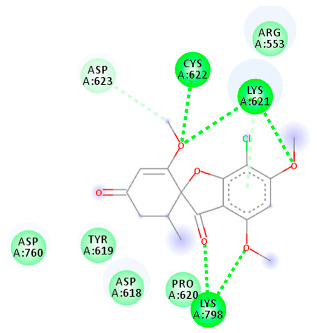
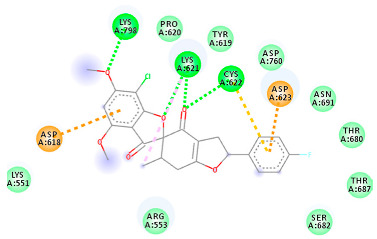
Molecular docking details of the top compounds selected for MD analysis. PubChem ID compound (first column), highlighted in red, blue, orange, and green, to indicate their interactions with M protease, ACE2, RdRp, and RBD, respectively. The dotted lines in the ligand interaction diagram (last column) show the key interacting residues the of targeted protein and ligand structure. Moreover, the green, light green, orange, pink, blue, and red dotted lines show conventional hydrogen bonds, van der Waals, cation–anion, pi-alkyl, halogen, and unfavorable acceptor–acceptor interactions, respectively.
| PubChem ID | Binding Energy (kcal/mol) | H-Bonds | Hydrophobic Interactions | Lipinski Analysis | Ligand Interaction Diagram | |
|---|---|---|---|---|---|---|
|
441140
(M1) |
−6.80 | HIS 41 GLY 143 CYS 145 GLU 166 |
MET 49 MET 165 ARG 188 |
Molecular weight Lipophilicity H bond donor H bond acceptor Violations Lipinski Hepatotoxicity |
352.7 2.95 0 6 0 Yes No |
|
| 142550537 (M7) | −7.58 | HIS 41 GLY 143 GLU 166 THR 190 GLN 192 |
MET 165 PRO 168 ARG 188 GLN 189 THR 190 GLN 192 |
Molecular weight Lipophilicity H bond donor H bond acceptor Violations Lipinski Hepatotoxicity |
506.7 2.59 0 13 2 Yes No |
|
| 144564153 (M9) | −9.49 | MET 165 GLU 166 |
CYS 44 MET 49 LEU 141 CYS 145 MET 165 |
Molecular weight Lipophilicity H bond donor H bond acceptor Violations Lipinski Hepatotoxicity |
484.9 3.61 1 7 0 Yes No |
|
| 441140 (A7) | −5.2 | HIS 401 ARG 514 |
HIS 378 GLU 398 HIS 401 |
Molecular weight Lipophilicity H bond donor H bond acceptor Violations Lipinski Hepatotoxicity |
352.7 2.95 0 6 0 Yes No |
|
| 46844082 (A3) | −8.44 | ALA 348 ASN 394 ARG 514 |
GLU 375 HIS 378 HIS 401 GLU 402 |
Molecular weight Lipophilicity H bond donor H bond acceptor Violations Lipinski Hepatotoxicity |
641.1 5.51 0 8 2 Yes No |
|
| 132286359 (A9) | −6.51 | ASP 382 GLY 395 PHE 400 HIS 401 GLU 402 ARG 514 |
GLU 375 HIS 378 HIS 401 GLU 402 |
Molecular weight Lipophilicity H bond donor H bond acceptor Violations Lipinski Hepatotoxicity |
551.3 3.85 2 9 1 Yes No |
|
|
441140
(Rd7) |
−5.62 | LYS 621 CYS 622 LYS 798 |
ASP 623 | Molecular weight Lipophilicity H bond donor H bond acceptor Violations Lipinski Hepatotoxicity |
352.7 2.95 0 6 0 Yes No |
|
| 73331209 (Rd2) | −7.21 | LYS 621 CYS 622 LYS 798 |
ASP 618 LYS 621 CYS 622 ASP 623 |
Molecular weight Lipophilicity H bond donor H bond acceptor Violations Lipinski Hepatotoxicity |
458.8 3.75 0 7 0 Yes No |
|
| 118254151 (Rd6) | −6.61 | LYS 621 CYS 622 LYS 798 |
ASP 618 LYS 621 CYS 622 ASP 623 ASN 691 |
Molecular weight Lipophilicity H bond donor H bond acceptor Violations Lipinski Hepatotoxicity |
458.8 3.73 0 7 0 Yes No |
|
|
441140
(Rb1) |
−5.3 | GLU 471 | LYS 458 GLU 471 |
Molecular weight Lipophilicity H bond donor H bond acceptor Violations Lipinski Hepatotoxicity |
352.7 2.95 0 6 0 Yes No |
|
| 56950846 (Rb3) | −7.21 | ARG 457 LYS 458 SER 469 |
ARG 457 LYS 458 GLU 471 PRO 491 |
Molecular weight Lipophilicity H bond donor H bond acceptor Violations Lipinski Hepatotoxicity |
428.8 3.6 0 6 0 Yes No |
|
| 125429633 (Rb9) | −6.5 | ARG 457 LYS 458 SER 459 GLU 471 ILE 472 |
ARG 457 LYS 458 GLU 471 PRO 491 |
Molecular weight Lipophilicity H bond donor H bond acceptor Violations Lipinski Hepatotoxicity |
516.9 3.45 2 9 1 Yes No |
|
