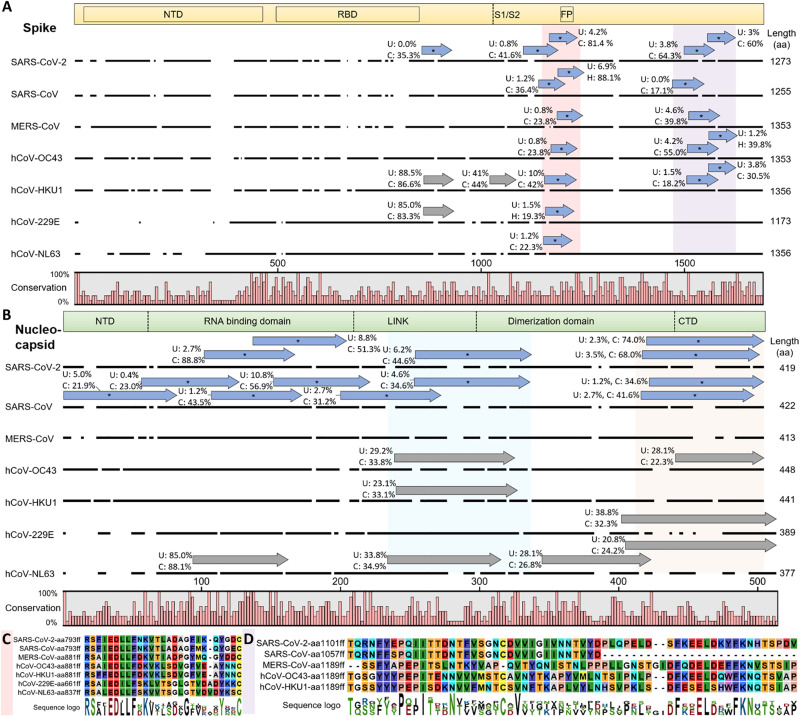
Fig. 3. hCoV cross-reactive antibody responses.
Cross-reactive and selective antibody binding of SARS-CoV-2 peptides and other hCoVs clusters in similar regions of the spike (A) and nucleocapsid (B) protein, with shared motifs of bound spike peptides highlighted (C and D). (A and B) Alignments of S and N proteins of all hCoVs. The dark line next to the strain identifier represents the protein sequence indicating gaps in the consensus alignment. Peptides bound at significantly different percentages (chi-squared/Kolmogorov-Smirnov tests and passing FDR correction; see data file S3) between unexposed (“U”) individuals or recovered patients with COVID-19 (“C”) are shown as blue arrows above the corresponding protein sequence and marked with an asterisk. The abundance of binding in U and C is indicated as percentages written next to the peptides. Gray arrows indicate similar recognition in >20% of unexposed individuals and patients with COVID-19. For SARS-CoV-2 only, peptides of the reference genome are included (variants not shown but listed in data file S3). The domain structure on top of each panel is based on SARS-CoV-2 S protein (68)/N protein (69), positions in other hCoVs shift along the alignment. Because of the different lengths of S and N proteins, the two panels are not drawn at the same scale. (C and D) Motifs from alignments of bound spike peptides in the regions marked in light red (C) and light purple (D) in (A). See fig. S3 (A and B) for full alignments of the peptides and details. Alignments of the nucleocapsid regions marked in light blue and light orange are due to space constraints shown in fig. S3 (C and D). aa, amino acid.