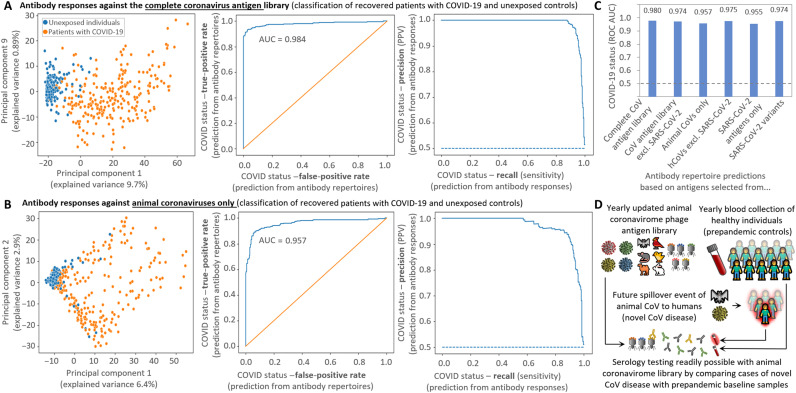
Fig. 7. Machine learning based diagnostics.
Antibody responses against antigens of aCoVs allow accurate identification of recovered patients with COVID-19 (A to C) suggesting a possible diagnostic strategy for the early stage of future pandemics (D). (A and B) Classification of recovered patients with COVID-19 (n = 269) and unexposed controls (n = 260) from antibody responses against all antigens of the CoV library (A) and only from the animal coronavirome (B). PCA on fold changes of antibody responses separates between hCoV antibody responses of unexposed individuals and recovered patients with COVID-19 (left) and a machine learning predictor accurately identifies infected individuals illustrated by receiver operating characteristic (ROC) curve (middle) and PR curve (right). (C) AUC of the ROC curve of predictions with subgroups of antigens of the CoV antigen library. All predictions [including (A) and (B)] were performed with gradient boosting decision trees [XGBoost classifier (70)] with leave-one-out cross-validation from CoV peptides bound in >5% individuals. The aCoVs used for these predictions are limited to RefSeq strains (deposited in 2018), whereas a bat CoV related to SARS-CoV-2 (24) was excluded (data file S1). (D) Creating regularly updated antigen libraries representing the animal coronavirome alongside the collection of baseline samples of healthy individuals can provide the basis for a serological assay to readily respond in case of a novel zoonotic transmission event.