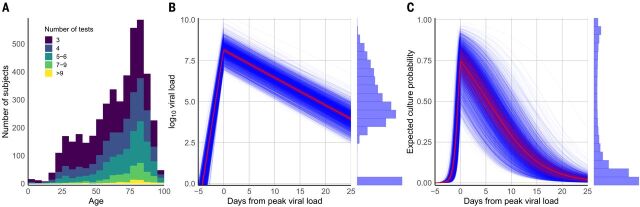
Fig. 4. Viral load and estimated infectious virus shedding time series.
Of 25,381 positive subjects, 4344 had three or more RT-PCR test results available, and these were used in a viral load time-series analysis. Subjects with only one result cannot be placed in time because of inherent ambiguity (given that the model has both an increasing and a decreasing phase), and those with only two test results are excluded from the time-series analysis because of insufficient data for temporal placement (their number of data points is less than the number of model parameters being estimated). (A) Number of subjects with three or more RT-PCR test results available, at least two of which were positive, according to age. (B) Estimated time course of viral load for 18,136 RT-PCR results from the 4344 subjects with at least three RT-PCR results. Blue lines are expected complete time courses for individual cases. The sample mean is shown in red, with its 90% credible interval as a shaded area. The histogram at right shows the distribution of all observed viral loads. The histogram values at zero correspond to the initial and trailing negative tests in subject timelines. Figure S8 shows raw viral load time series, per subject and split by number of RT-PCR tests. (C) Estimated time course of positive cell culture probability, calculated by applying the results shown in Fig. 2C to the estimated viral load time courses in (B). Blue lines are expected time courses for individual subjects. The sample average is shown in red, with its 90% credible interval as a shaded area. The histogram at right shows the distribution of culture probabilities in the sample and was obtained by applying the curve in Fig. 2C to the data in the histogram in (B).