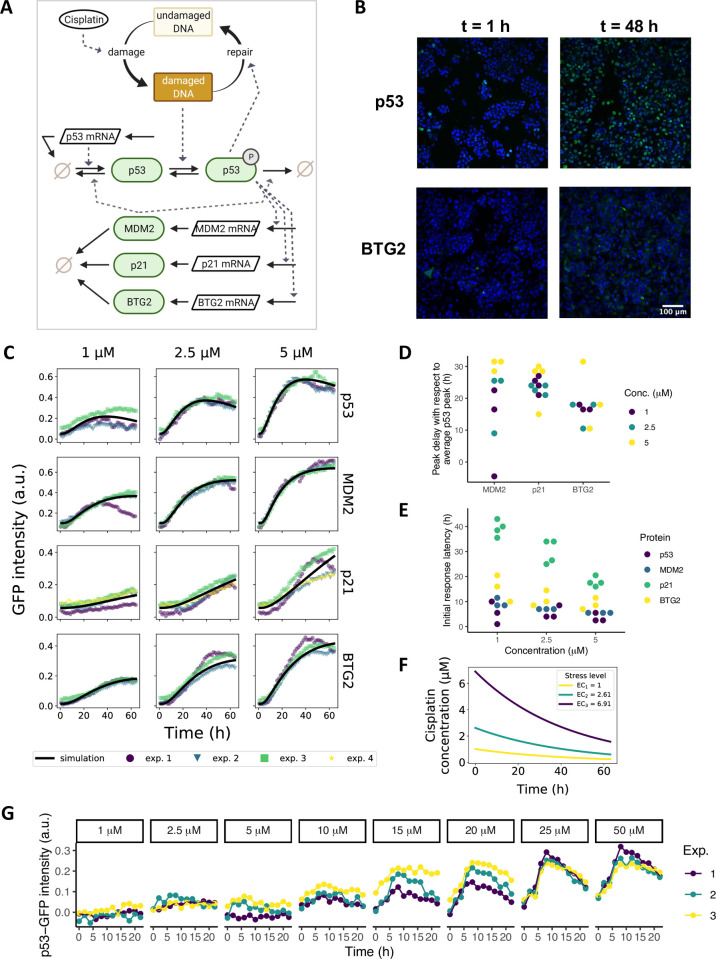
Fig 3. ODE modeling of cisplatin-induced p53 signaling in HepG2 cells calibrated on live cell confocal imaging data.
(A) Graphical representation of the model. Cisplatin causes DNA damage, which triggers phosphorylation of p53. Phosphorylated p53 induces expression of its downstream targets MDM2, p21 and BTG2. (B) Example images of nuclear GFP expression in p53, and cytoplasmic GFP expression in BTG2 HepG2 BAC-GFP reporter cells at 1 and 48 hours. Blue, Hoechst-stained nuclei; green, GFP signal. (C) Mean of single-cell protein expression data measured over a 65-hour period after 1, 2.5 and 5 μM cisplatin exposure. Experimental replicates (exp. 1–4) are shown in different colors. The model simulation after parameter calibration is shown as a black solid line. (D) Delay in peak MDM2, p21 and BTG2 expression levels with respect to maximum p53 expression. (E) Response latency of MDM2, p21 and BTG2 for different cisplatin concentrations. (F) Model-predicted effective cisplatin concentrations (ECi) over time. Corresponding applied concentrations are 1, 2.5 and 5 μM for EC1, EC2 and EC3, respectively. (G) Induction of p53-GFP in three replicates (Exp. 1–3) after exposure of non-treated HepG2 cells to conditioned medium collected from cells previously exposed to cisplatin for 72 hours.