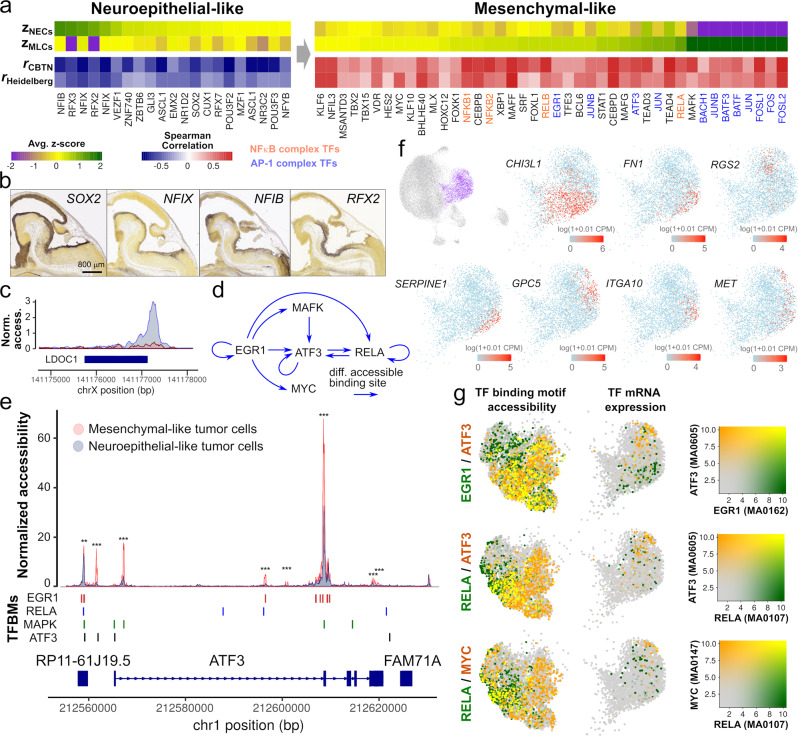
Fig. 4. The transition of posterior fossa ependymoma tumor cells into a mesenchymal-like state involves the inhibition of neuroepithelial transcription factors and the activation of the NFκB and AP-1 complexes.
a Differentially expressed transcription factors in the neuroepithelial- or mesenchymal-like tumor cell populations that have differentially accessible binding motifs across the EMT (FDR < 0.1). The heatmap shows the transcription factor binding motif score in the neuroepithelial (zNECs) and mesenchymal-like (zMLCs) cell tumor cell populations according to the single-nucleus ATAC-seq data, and the Spearman’s correlation coefficient between the inferred abundance of MLCs and the expression level of the transcription factor in the CBTN (rCBTN) and Heidelberg (rHeidelberg) bulk RNA-seq cohorts. Transcription factors that are part of the NFκB and AP-1 complexes are indicated in orange and blue, respectively. b Whole-mount RNA in situ hybridization of sagittal sections of E13.5 mouse embryos, showing the expression of neuroepithelial transcription factors in the neuroepithelum of the brainstem and isthmic organizer (Image credit: Allen Institute). c Normalized chromatin accessibility profile at the LDOC1 locus for neuroepithelial-like and MLC populations, showing the inaccessibility of the promoter in MLCs. d Inferred regulatory network between some members of the MAF/BACH, NFκB, and AP-1 complexes. Arrows from one transcription factor into another indicate the presence of at least one differentially accessible binding motif of the first transcription factor in the gene locus of the second transcription factor (Fisher’s exact test, FDR ). This analysis indicates ATF3 integrates signals from the AP-1, ERK, MAF/BACH, and NFκB signaling pathways. e Normalized chromatin accessibility at the ATF3 gene locus for neuroepithelial- and mesenchymal-like tumor cells, where the binding sites of EGR1, RELA, MAPK, and ATF3 are indicated. Transcription factor binding sites are differentially accessible between the neurepithelial- and mesenchymal-like cell populations. (Two-sided Fisher’s exact test; *FDR , **FDR , ***FDR ). Numeric q values are provided in the Source Data file. f The part of the UMAP representation corresponding to the MLC population is colored by the expression level of several genes with significantly heterogeneous expression within that cluster (Laplacian score, FDR < 0.01) based on a spectral graph method. g The part of the single-nucleus ATAC-seq and RNA-seq UMAPs that corresponds to the MLC population is colored by the transcription factor binding motif accessibility score and the gene expression level of RELA, ATF3, EGR1, and MYC transcription factors. In the figure, a substantial amount of heterogeneity is observed within the MLC population, and the relative patterns of expression and motif accessibility are consistent between the single-nucleus RNA-seq and ATAC-seq representations. MLCs mesenchymal-like tumor cells, NECs neuroepithelial-like tumor cells, TFBMs transcription factor binding motifs.