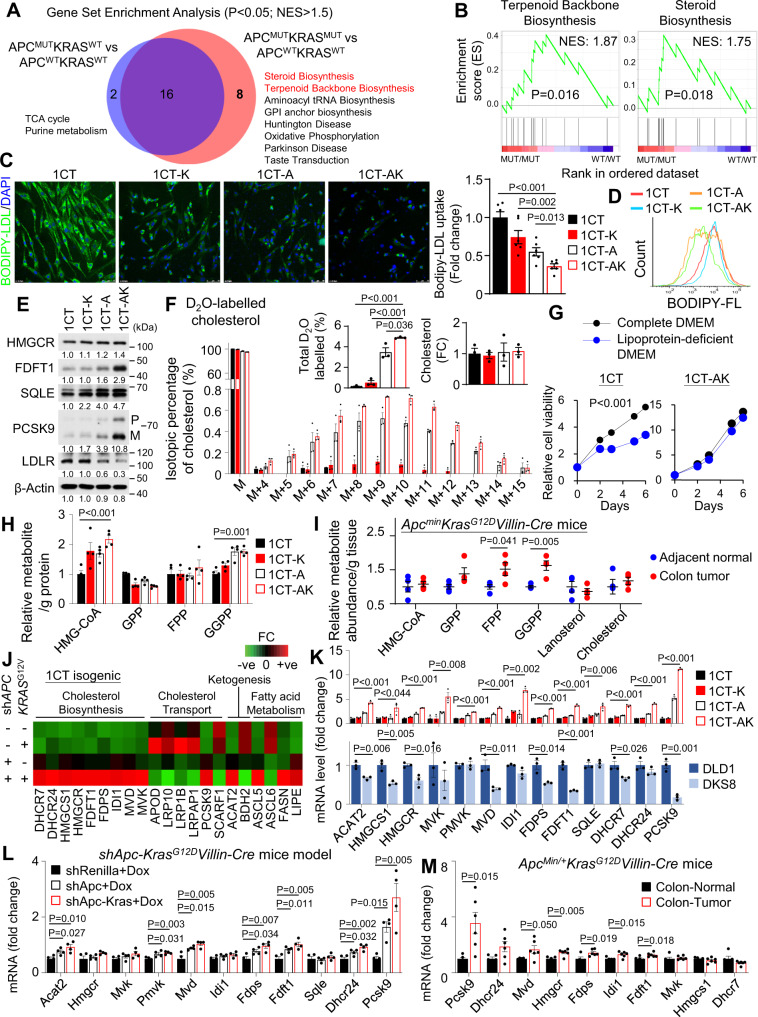
Fig. 1. Cholesterol homeostasis is shifted from cellular uptake to de novo biosynthesis in APC/KRAS-mutant CRC leading to increase geranylgeranyl pyrophosphate (GGPP) levels.
A Gene Set Enrichment Analysis (GSEA) of enriched KEGG pathways in APC-mutant (N = 142), APC/KRAS-mutant CRC (N = 134) as compared to wildtype counterparts (N = 50) from the Cancer Genome Atlas (TCGA) colon and rectal cancer (COADREAD) cohort. Permutation-based p-value. B Terpenoid Backbone Biosynthesis pathway and Steroid Biosynthesis pathway are enriched in APC/KRAS-mutant CRC cases from TCGA cohort. Permutation based p-value. C Fluorescence microscopy of cellular uptake of fluorescent labelled LDL (BODIPY-LDL) in 1CT isogenic cell lines (1CT-K: 1CT-KRASG12V, 1CT-A: 1CT-shAPC, 1CT-AK: 1CT-shAPC-KRASG12V). Reduced LDL uptake was observed in 1CT-AK cells (n = 6). D Flow cytometry of intracellular fluorescence after incubation with BODIPY-LDL. E Western blot confirmed the up-regulated protein expression of PCSK9 and cholesterol biosynthesis genes in 1CT-AK cells; LDLR was inhibited. F D2O stable isotope labeling (48 h) of cholesterol and LC-MS analysis of intracellular cholesterol in 1CT isogenic cells, revealing increased de novo cholesterol biosynthesis in 1CT-A and 1CT-AK cells, while having no effect of total cholesterol level (n = 3). G Cell viability of 1CT-AK cells were not sensitive to lipoprotein depletion in culture medium. Cell growth curve of 1CT and 1CT-AK cells in complete or lipoprotein-depleted medium (n = 3). H LC-MS of cholesterol pathway intermediates in 1CT isogenic cells. I LC-MS of cholesterol pathway intermediates in CRC and adjacent normal tissues from 7-weeks-old Apcmin/+KrasG12D/+Villin-Cre mice (n = 4). J PCR array (Human lipoprotein signaling & cholesterol metabolism; and Human fatty acid metabolism) revealed that KRASG12V plus shAPC increased mRNA expression of cholesterol biosynthesis genes. K qPCR validated the increased mRNA expression of PCSK9 and cholesterol biosynthesis genes in 1CT-AK cells compared to 1CT, 1CT-K, and 1CT-A cells, whilst knockout of mutant KRAS in DLD1 cells (i.e., DKS8) exerted an opposite effect (n = 3). L PCSK9 and cholesterol biosynthesis genes were up-regulated in the shApc-KrasG12D-Villin-Cre mice colon (n = 4) compared to shRen controls (n = 4) and shApc-Villin-Cre mice (n = 4). M mRNA levels of PCSK9 and cholesterol biosynthesis genes in ApcMin/+KrasG12DVillin-Cre mice tumors (n = 6) compared to adjacent normal tissues (n = 5). Data shown are means of biological replicates; ± SEM (C, F–I, K–M). Two-tailed Student’s t-test (C, F, H, I, K–M). Two-tailed two-way ANOVA (G). Source data are provided as a Source Data file.