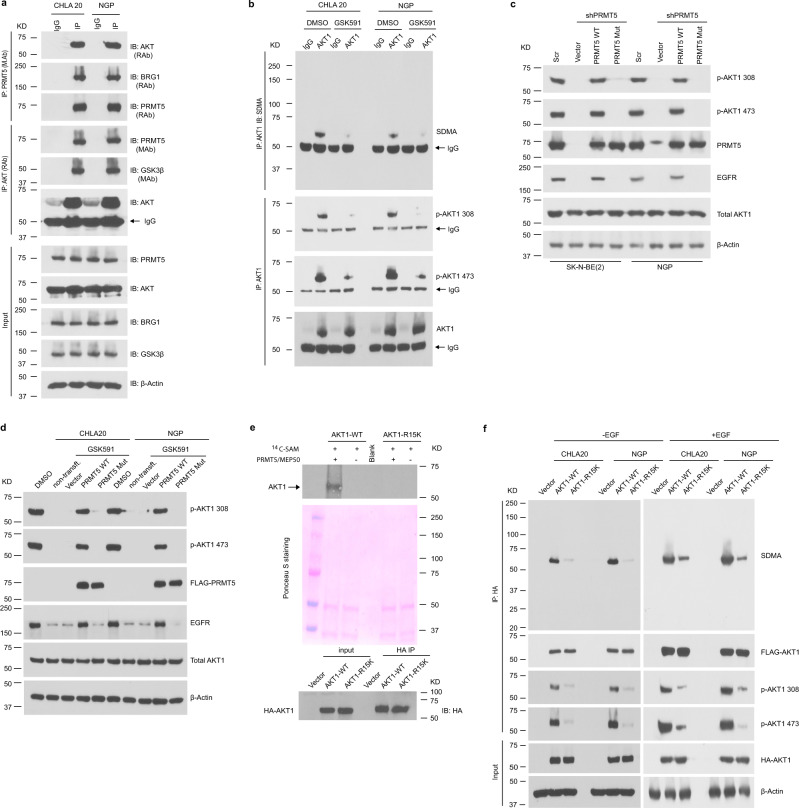
Fig. 4. PRMT5 methylates AKT.
a PRMT5/AKT interaction captured by co-immunoprecipitation (co-IP). The lysate was immunoprecipitated with anti-PRMT5 antibody followed by immunoblotting with anti-AKT antibody in CHLA20 and NGP cells (top), and the reciprocal co-immunoprecipitation was shown in the middle. BRG1 and bait protein PRMT5 were shown as positive controls for the PRMT5 IP, whereas GSK3β and bait protein AKT served as positive controls for AKT IP. The input was used as internal controls (bottom). b Immunoprecipitation of AKT1 followed by a Western blotting analysis of symmetric dimethylarginine (SDMA) of AKT1, phosphorylation of AKT1 on Thr308 and Ser473 in DMSO or GSK591-treated CHLA20 and NGP cells. c AKT1 phosphorylation was detected by immunoblotting in a scramble or PRMT5 knockdown cells when forced expressing a wild-type PRMT5 or an enzymatic deficient form of PRMT5. d Analysis of AKT1 phosphorylation in DMSO or GSK591-treated CHLA20 and NGP cells expressing PRMT5 wild type or enzyme dead mutant. e In vitro methylation assay showing the methylation of AKT1 wild type and R15K mutant by recombinant PRMT5/MEP50 (top), Ponceau S staining of the membrane showing equal loading of each sample (middle), and Western blotting analysis showing an equal amount of HA-tagged proteins pulled down by anti-HA beads (bottom). f Analysis of SDMA and phosphorylation of AKT1 wild type or R15K mutant in CHLA20 and NGP cells with (right) or without (left) EGF stimulation. Cells were transfected with AKT1 wild type or R15K mutant. In the case of EGF stimulation, 24 h post-transfection, cells were serum-starved overnight and then treated with 10 ng/mL EGF for 15 min before harvest. a–d, f Representative results from three independent experiments. e Representative results from two independent biological samples. Uncropped immunoblots are provided in the Source Data file.