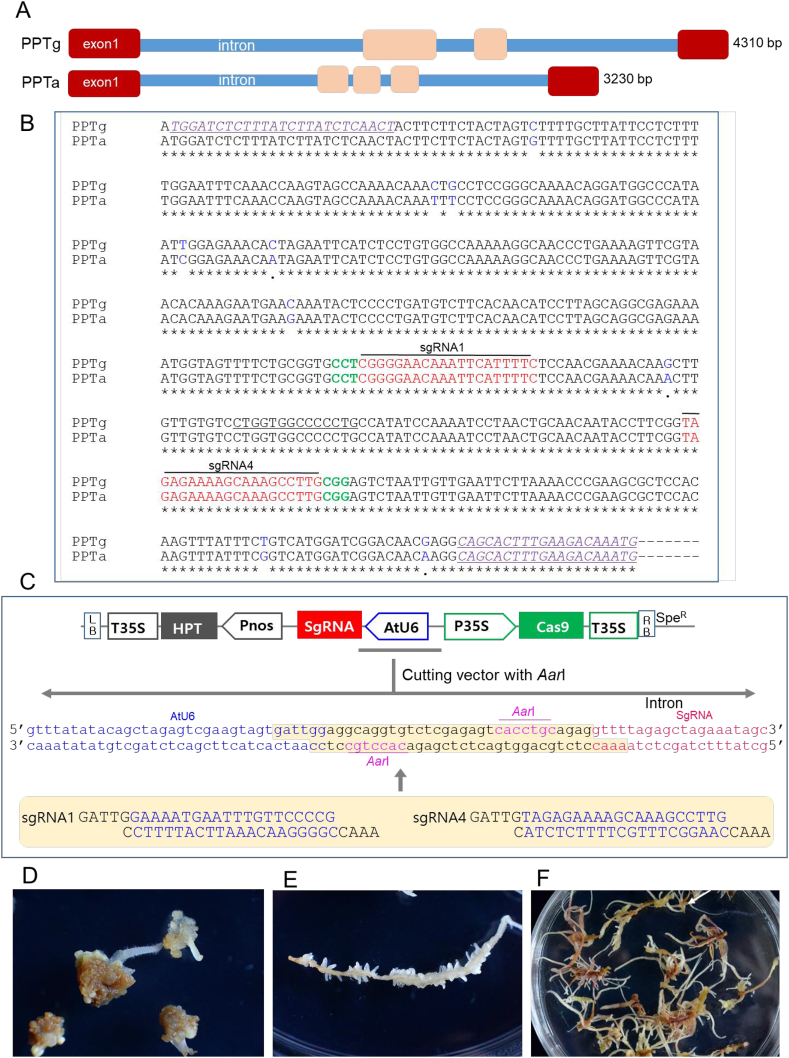
Fig. 2.
Selection of target sites in the PPT synthase gene sequence, CRISPR/Cas9 binary vector construction, and ginseng transformation. (A) Intron and exon structure of PPTg and PPTg sequences. The first exon regions (exon 1) with 473 bp containing start codon in both PPTg and PPTg were highly homologous. (B) Locations of two target sites (sgRNA1 and sgRNA4) for CRISPR/Cas9-mediated target mutagenesis in the N-terminal site of PPTa and PPTg exon sequences. sgRNA1 (designed using the reverse-frame sequence) and sgRNA4 are marked in red, PAM sequences are marked in green, and primers used for sequencing are underlined and purple. Nine blue letters are single nucleotide polymorphisms between the PPTa and PPTg sequences. (C) Schematic representation of the CRISPR/Cas9 binary vector used for ginseng transformation. Arabidopsis thaliana promoters and terminators drive the expression of gRNA1 (AtU6–26p and AtU6–26t) and gRNA4 (AtU6–29p and AtU6–29t). The caulifower mosaic virus promoter (CaMV 35S) drives the expression of the Cas9 gene. The two sgRNAs were inserted into vectors using AarI enzyme sites. (D) Formation of new adventitious roots on the surfaces of hygromycin-resistant callus derived from root segments on medium with 2 mg/l IBA and 20 mg/l hygromycin. (E) Proliferation of an excised single root by induction of lateral root formation on medium with 2 mg/l IBA. (F) Proliferation of adventitious root masses from a single root for further analysis.