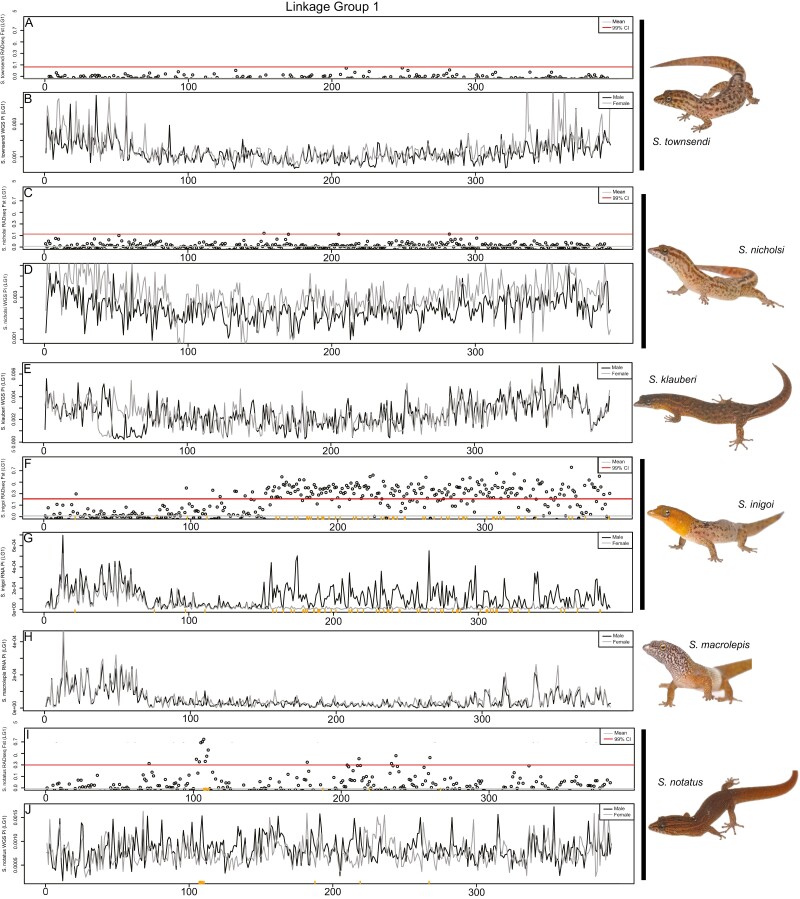
Figure 5.
Comparative genomics of the S. notatus sex chromosome (LG1) across multiple Sphaerodactylus species in 500kb windows. (A and B) S. townsendi M/F FST values (RADseq) and M and F nucleotide diversity (WGS), respectively; (C and D) S. nicholsi M/F FST values (RADseq) and M and F nucleotide diversity (WGS), respectively; (E) S. klauberi M and F nucleotide diversity (WGS); (F and G) S. inigoi M/F FST values (RADseq) and M and F nucleotide diversity (RNAseq), respectively; (H) S. macrolepis M and F nucleotide diversity (RNAseq); (I and J) S. notatus M/F FST values (RADseq) and M and F nucleotide diversity (WGS), respectively. Sex-specific RADtags mapped to S. inigoi (F and G) and S. notatus (I and J) along the X axis (orange ticks). Note: slight shifts on the X-axis are due to the differences in programs used to calculate values, i.e., WGS used pixy, while RADseq and RNAseq used vcftools.