Fig. 1.
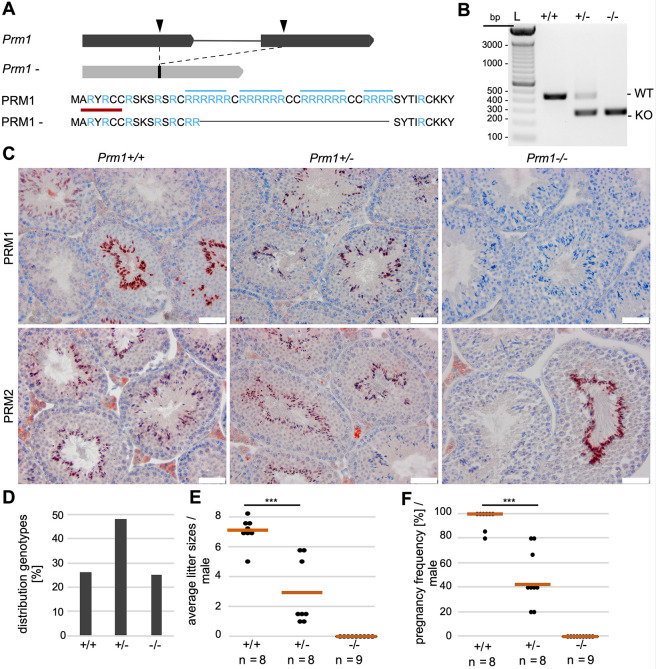
Establishment of Prm1-deficient mice and fertility analysis. (A) Graphical representation of CRISPR-Cas9-mediated gene editing of the Prm1 locus. Two sgRNAs were used (black arrowheads), targeting the Prm1 coding sequence in exon 1 and exon 2, respectively. A 167 bp in-frame deletion was generated, leading to loss of crucial arginine-rich DNA-binding sites (marked in blue). The epitope of the anti-PRM1 antibodies used in (C) is marked in red. (B) Agarose gel of genotyping PCR of Prm1+/+, Prm1+/− and Prm1−/− mice. Amplification of WT Prm1 or the Prm1− allele generated products of 437 bp or 270 bp, respectively. (C) IHC staining against PRM1 and PRM2 on Bouin-fixed, paraffin-embedded testis sections of Prm1+/+, Prm1+/− and Prm1−/− mice counterstained with Hematoxylin. (D) Mendelian distribution of genotypes (n=10 litters) from crossings of Prm1+/− males and females. (E) Scatter plot of mean litter sizes monitored per male after mating with female WT C57BL/6J mice. The mean litter size per genotype is indicated by vermillion lines. (F) Pregnancy frequency (%) per male after mating with female WT C57BL/6J mice. n=number of males used; data are means and were analyzed using a two-tailed, unpaired Student's t-test (***P<0.001). Scale bars: 50 µm. L=ladder.