Fig. 1. The INDUCE-seq workflow.
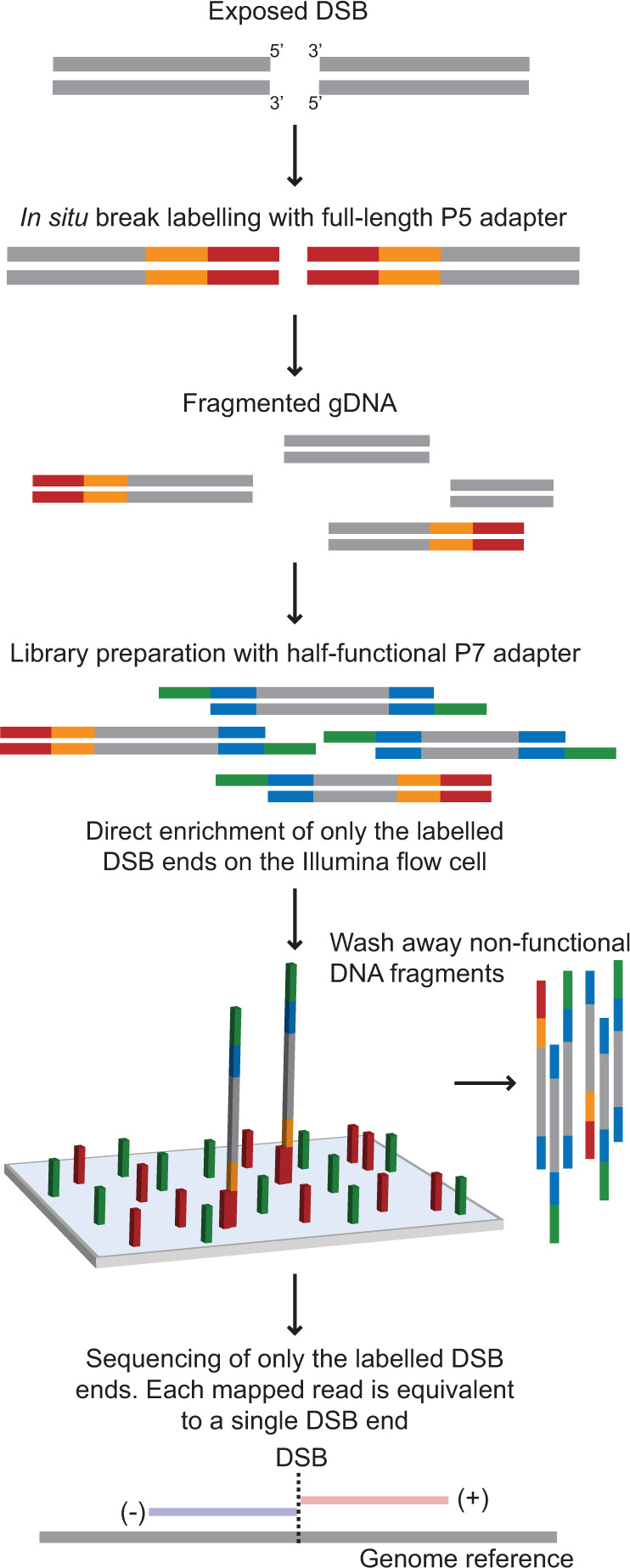
In situ break labelling in fixed and permeabilised cells is performed by ligating a full-length, chemically modified P5 sequencing adapter to end-prepared DSBs. Genomic DNA is then extracted, fragmented, end-prepared and ligated using a chemically modified half-functional P7 adapter. Resulting DNA libraries contain a mixture of functional DSB-labelled fragments (P5:P7) and non-functional genomic DNA fragments (P7:P7). Subsequent illumina sequencing of INDUCE-seq libraries enriches for DNA-labelled fragments and eliminates all other non-functional DNA. As the INDUCE-seq library preparation is PCR-free, each sequencing read obtained is equivalent to a single labelled DSB-end from a cell.
