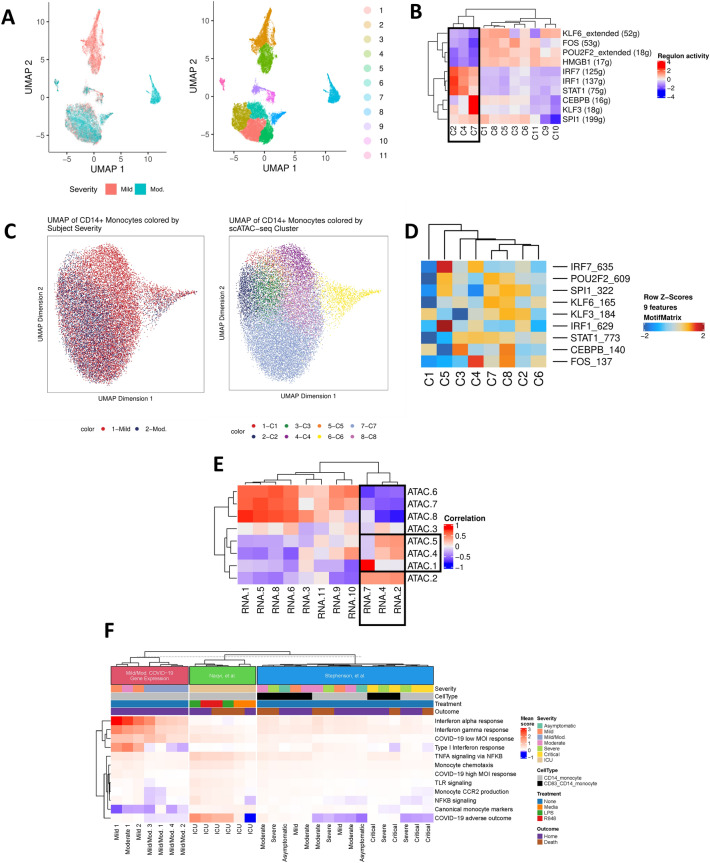
Figure 5.
CD14+ monocytes undergo extensive chromatin remodeling prior to seroconversion and harbor severity-specific epigenetic biomarkers. (A) scRNA-seq UMAP of all CD14+ monocytes collected from mild and moderate IgG− subjects. UMAP colored by disease severity (left) and cluster number (right). (B) Heat map depiction of regulon activity computed for each scRNA-seq cluster using SCENIC. Activity of the top 10 regulons (right) is plotted for each cluster of cells from the UMAP (bottom). The black box indicates clusters of interest. (C) scATAC-seq UMAP of all CD14+ monocytes collected from IgG− mild and moderate subjects. UMAP colored by disease severity (left) and cluster number (right). (D) Heat map depiction of transcription factor motif enrichment of identified regulons (right) plotted for each scATAC-seq cluster (bottom). (E) Correlation plot using regulon activity to link clusters between scRNA-seq (columns) and scATAC-seq (rows). Black box indicates clusters of interest. (F) Enrichment of gene expression in monocyte-related pathways for CD14+ monocyte subsets from IgG− mild and moderate subjects. Pathway scores were computed for cell subsets from two published datasets for comparison. Hierarchical clustering was applied to samples within each dataset.