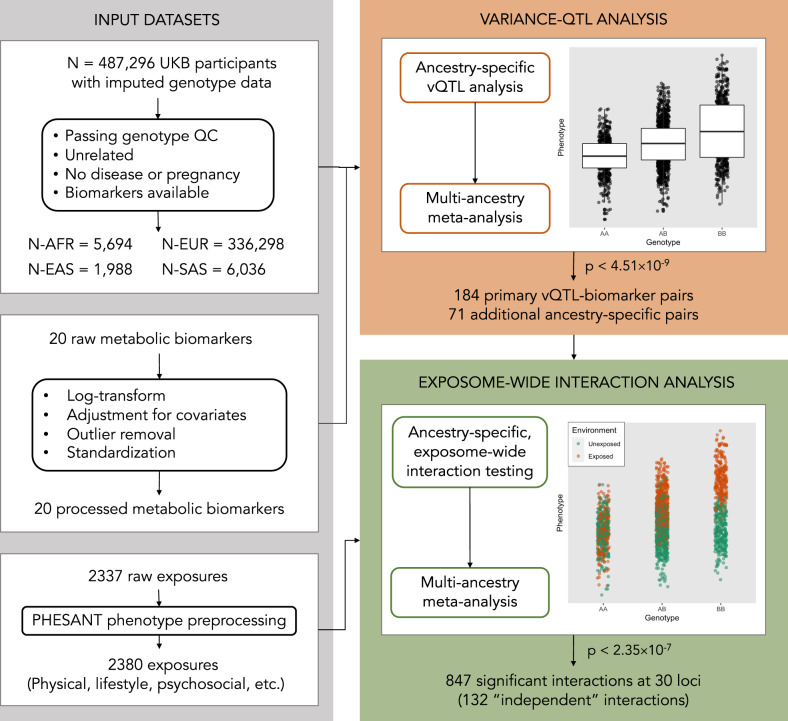
Fig. 1. Analysis workflow.
Unrelated individuals without major disease from any of four ancestry groups in the UK Biobank were included in the analysis. Twenty metabolic biomarkers were pre-processed, including log transformation, adjustment for biological and technical covariates, and outlier removal. vQTL analysis was performed genome-wide, separately in each ancestry group followed by a multi-ancestry meta-analysis. Significant variant-biomarker pairs were taken forward for GEI analysis, along with 2380 exposures pre-processed using the PHESANT tool. Finally, GEI tests were performed for each combination of variant-biomarker-exposure triplet, again using ancestry-stratified analysis followed by meta-analysis.