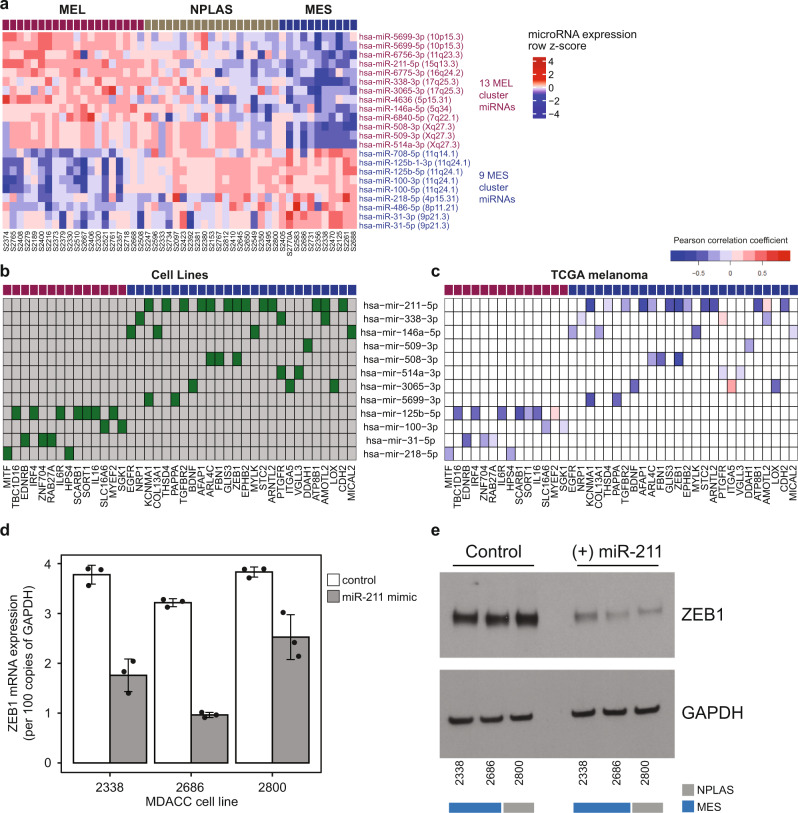
Fig. 5. Inverse associations between MEL and MES gene expression and mature strand miRs in cell lines are preserved in melanoma tumors.
a Heatmap depicting expression of miRs differentially expressed between MEL and MES early passage MDACC melanoma cell lines (n = 53). b Inverse Pearson correlations between abundances of miRs differentially expressed between MEL and MES samples, and the group of mRNAs confirmed by at least one experimentally validated and/or at least two in silico miR target databases to be targets of these miRs, in the early passage cell lines. More miRs targeting MES genes than MEL genes were identified; 8 active MES-targeting miRs (highly expressed in MEL samples) and 4 active MEL-targeting miRs (highly expressed in MES samples). c The majority of miR-mRNA associations identified in cell lines were also observed in TCGA melanoma tissue samples. d Effects of miR-211-5p overexpression in the cell lines indicated on ZEB1 mRNA expression. Bars depict mean + SD of three biological replicates under control (white bars) or miR-211-5p mimic (gray bars) treatment conditions. e Representative Western blot of ZEB1 protein expression in the cell lines indicated comparing control and miR-211-5p overexpression conditions. Source data are provided as a Source Data file.