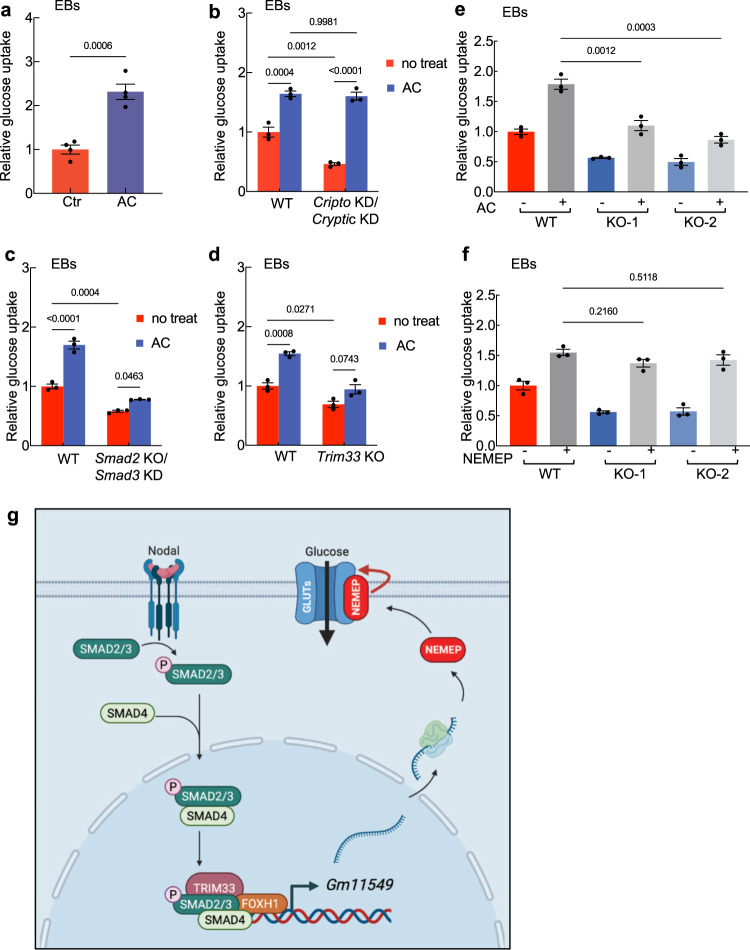
Fig. 7. Nodal signaling mediates glucose uptake during mesendoderm differentiation.
a Glucose uptake analysis in control and Activin A (AC)-treated for 2 h EBs. The values are normalized to the protein concentration (n = 4 biological independent samples). b Glucose uptake analysis in Activin A (AC)-treated for 2 h or untreated WT and Cripto KD/Cryptic KD EBs. The values are normalized to the protein concentration (n = 3 biological independent samples). c Glucose uptake analysis in Activin A (AC)-treated for 2 h or untreated WT and Smad2 KO/Smad3 KD EBs. The values are normalized to the protein concentration (n = 3 biological independent samples). d Glucose uptake analysis in Activin A (AC- treated for 2 h or untreated WT and Trim33 KO EBs). The values are normalized to the protein concentration (n = 3 biological independent samples). e Glucose uptake analysis of Activin A (AC)-treated for 2 h or untreated WT and Nemep KO EBs at day 3. The values are normalized to the protein concentration (n = 3 biological independent samples). f Glucose uptake analysis of NEMEP-FLAG overexpressing WT and Nemep KO EBs at day 3. The values are normalized to the protein concentration (n = 3 biological independent samples). g Working model for the role of NEMEP during mesendoderm differentiation (Created with BioRender.com). A highly conserved putative lncRNA Gm11549 is specifically induced by Nodal signaling during mesendoderm differentiation and it is essential for mesendoderm differentiation in mESCs. There is a “ORF” hidden in Gm11549 that codes a transmembrane micropeptide, we named NEMEP. NEMEP functions by interacting with GLUT1 and GLUT3, and augment the glucose uptake capacity of glucose transporter proteins to meet the energy needs during mesendoderm differentiation. a–f: Data are the mean ± S.E.M. P values were determined by unpaired two-tailed t-tests (a), one-way ANOVA (e, f) with Dunnett’s corrections, two-way ANOVA (b–d) with Sidak’s corrections, and data are representative of three independent experiments with similar results. Source data are provided as a Source Data file.