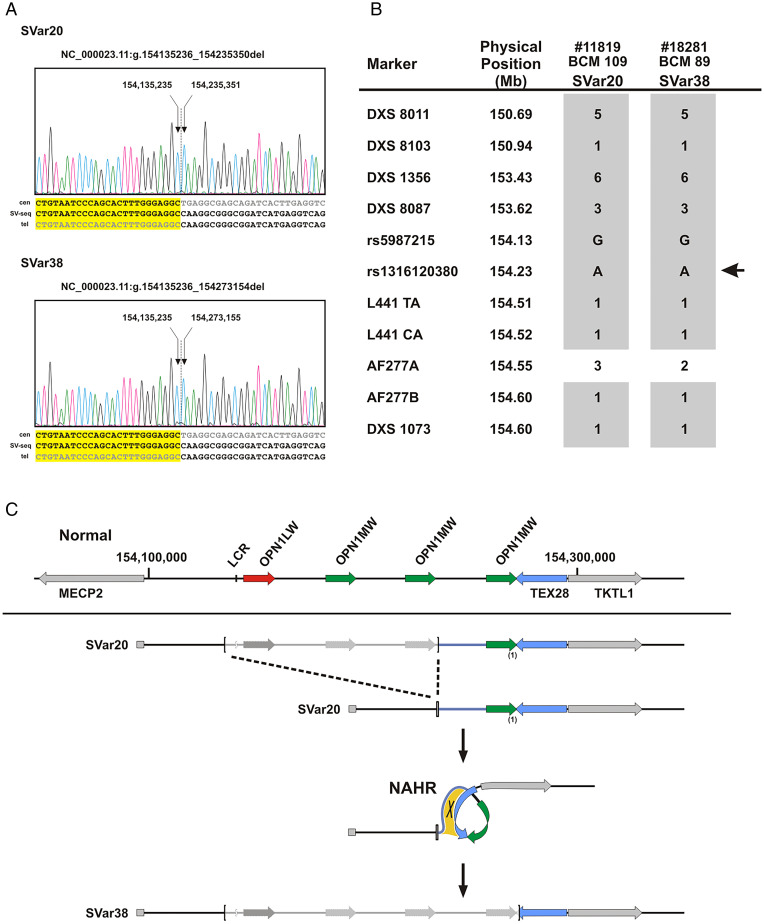
Fig. 3.
Identical centromeric breakpoints and sequence-conserved telomeric breakpoints in SVar20 and SVar38 support a single lineage intrachromosomal NAHR event. (A) SVar20 and SVar38 share identical centromeric breakpoints while telomeric breakpoints share the same breakpoint sequence but differ by the presence/absence of a single OPN1MW gene copy. (B) Patient #11819/BCM 109 carrying SVar20 and patient #18281/BCM 89 carrying SVar38 share the same marker haplotype at Xq28. Markers including nine microsatellites and two SNPs are ordered according to their physical position (top to bottom). The localization of the OPN1LW/OPN1MW gene cluster is indicated by the arrow on the right. Shared alleles (microsatellite alleles coded in numbers) are depicted as gray squares. (C) Proposed sequence of events linking SVar20 and SVar38. Deletion of the LCR and parts of the OPN1LW/OPN1MW gene cluster results in SVar20, which retains a single OPN1MW gene copy. Subsequently, SVar20 undergoes intrachromosomal NAHR through homologous sequences downstream of the OPN1MW gene copies (relevant area of sequence homology indicated by the yellow patch; note that the intergenic sequence between OPN1MW gene copies is homologous to large parts of the TEX28 gene) which results in the loss of the terminal OPN1MW gene copy as seen in SVar38. In comparison with the structure of the normal gene cluster (Top), the SV breakpoints are marked by brackets and deleted parts are indicated by lighter gray color.