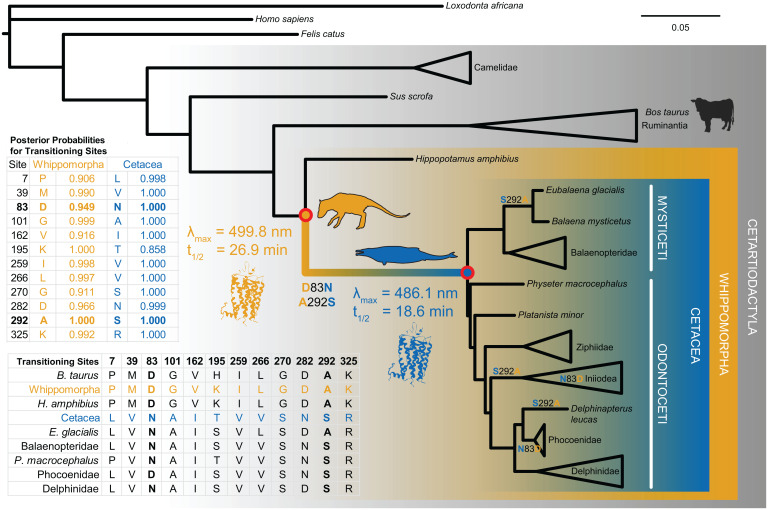
Fig. 1.
Phylogeny of cetartiodactyl rhodopsin coding sequences and reconstructed terrestrial-to-aquatic transitioning residues. We used codon-based likelihood models of evolution to infer the rhodopsin coding sequence at internal nodes of the tree (Cetacea and Whippomorpha). The reconstructed sequences had high posterior probabilities and revealed 12 transitioning residues. We then synthesized and expressed the ancestral genes in vitro via transfection into mammalian tissue culture cells for experimental tests of evolutionary hypotheses. We found significant shifts in both spectral tuning (λmax) and meta II decay (retinal release t1/2) along the transitional branch, caused by substitutions D83N and A292S. The tree topology shown is based on established species relationships, where branch lengths are proportional to the number of nucleotide substitutions per codon (see Materials and Methods and SI Appendix for further details).