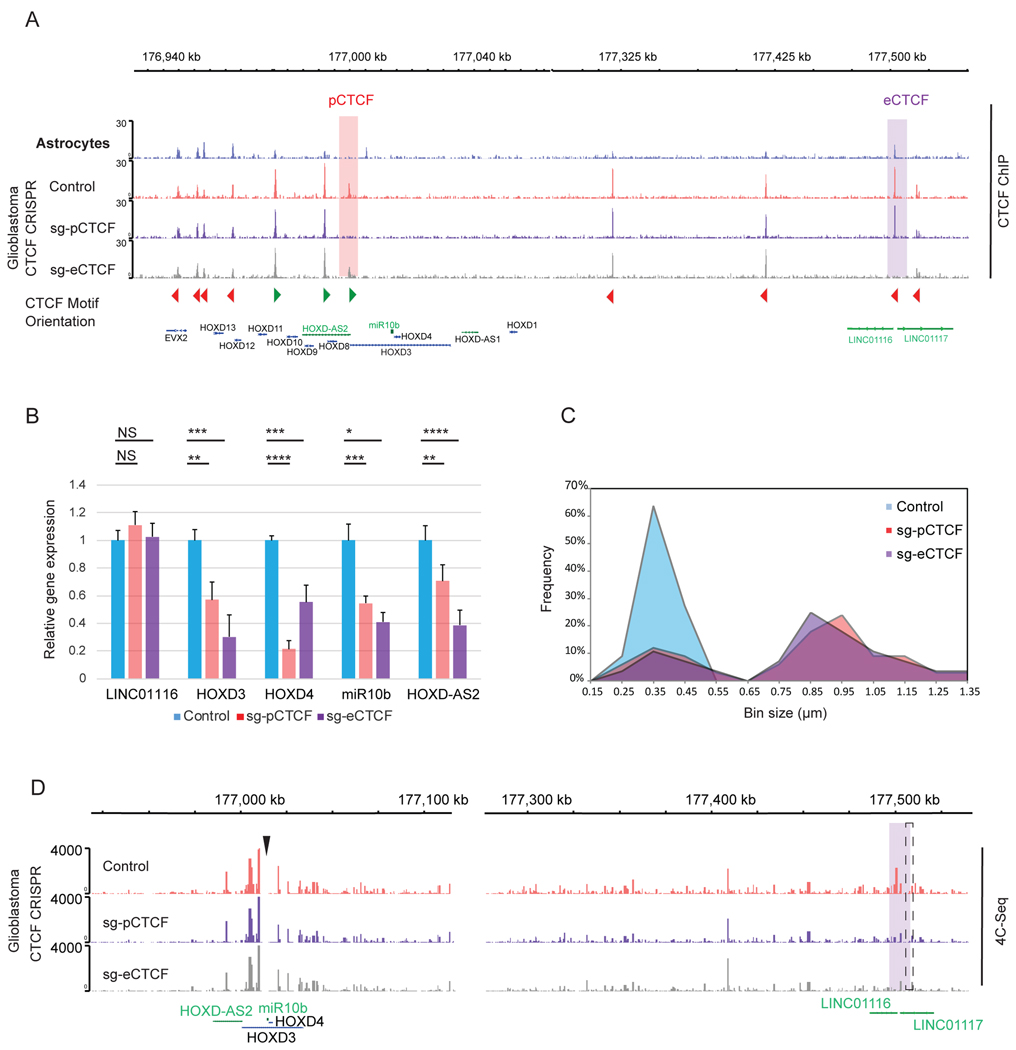
Figure 5. CRISPR-Cas9 editing of specific CTCF binding sites at the HOXD3/miR-10b promoter and enhancer recapitulates the effects of HOXD-AS2 and LINC01116 KD.
A. The CTCF binding sites in GBM and astrocytes, with representative visualization. Differential CTCF peaks in HOXD3/HOXD4/miR10b promoter (pCTCF) and LINC01116 enhancer (eCTCF) selected for editing, are highlighted in pink and purple, respectively. Two representative bottom tracks validate the loss of binding upon CRISPR-Cas9 editing of the corresponding sites (n=2).
B. CRISPR-Cas9 editing of pCTCF and eCTCF sites reduces expression of HOXD mRNAs and miR-10b in glioma cells. LN229 cells were treated with editing plasmids containing non-targeting Control sgRNA (blue), pCTCF (red) or eCTCF-targeting sgRNAs (purple), and gene expression was quantified by qRT-PCR (mean+SD, n=3).
C. CRISPR-Cas9 editing of pCTCF and eCTCF sites reduces the colocalization of HOXD-AS2 and LINC01116 transcripts. Frequency distribution of the distances between LINC01116 and HOXD-AS2 RNA are shown in control LN229 cells (Control sgRNA, blue), and cells edited with pCTCF sgRNA (red) or eCTCF sgRNA (purple). Axis X represents the bin intervals in μm, with indicated midpoints. The measurements were taken for 30 cells per condition.
D. CRISPR editing of either pCTCF or eCTCF reduces interactions between HOXD and LINC01116 enhancer. 4C-Seq was performed on LN229 with HOXD3/D4/miR10b bait (arrowhead). The major interaction, lost upon editing, is highlighted in purple. The representative track for each group is shown. Puncture line indicates the CRISPR-edited eCTCF region.