Figure 1 |. The definition of community-driven Microscopy Image Data Standards requires three complementary components and needs a flexible framework to manage complexity.
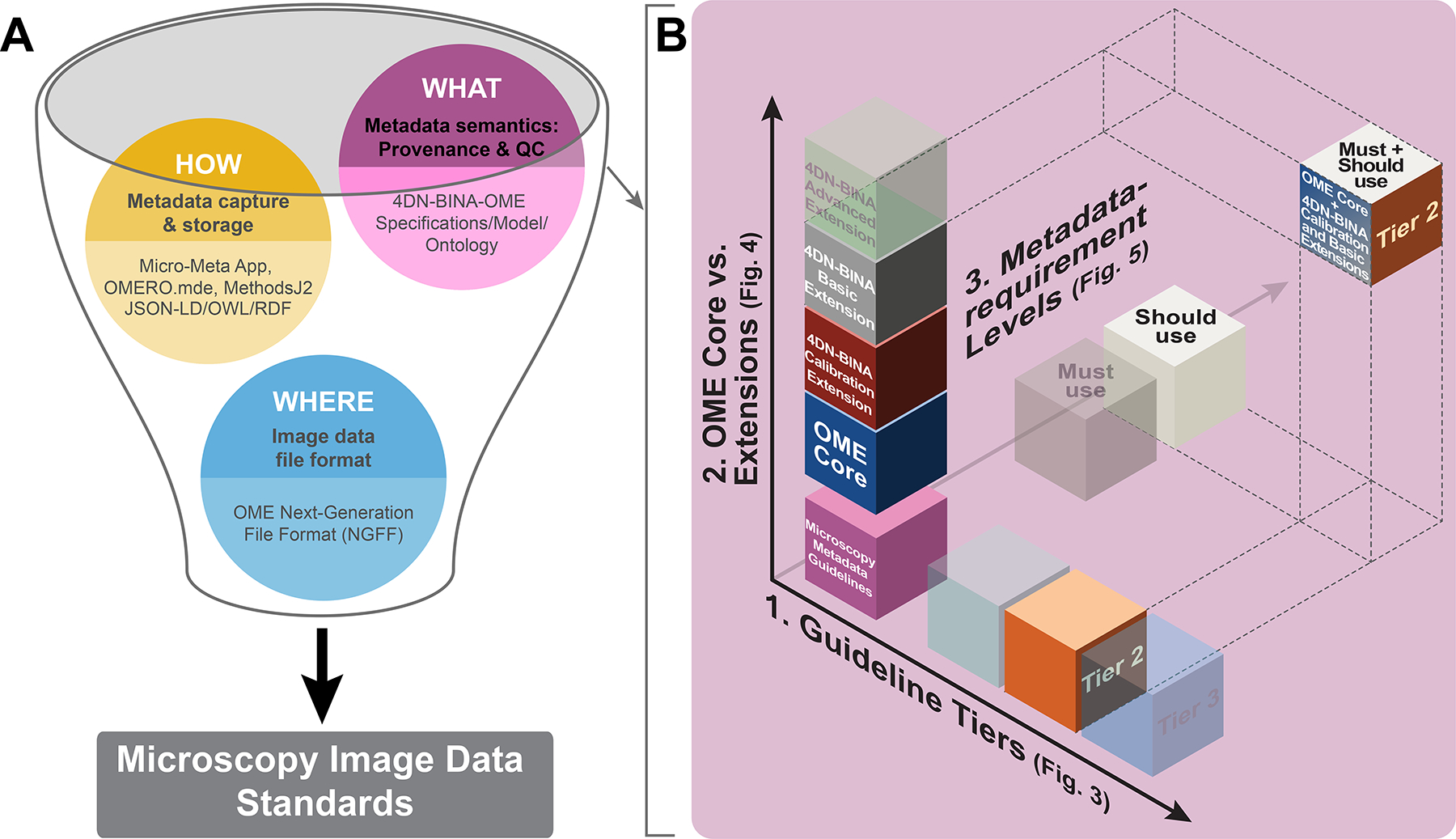
A) The establishment of community-driven Microscopy Image Data Standards requires development on three interrelated fronts: 1) Community-driven specifications for WHAT Microscopy Metadata information about an imaging experiment are essential for rigor, reproducibility, and reuse and should therefore be captured in Microscopy Metadata (magenta bubble); 2) Shared rules for HOW the (ideally) automated capture, representation and storage of Microscopy Metadata should be implemented in practice (yellow bubble). Last but not least, 3) Next-Generation File Formats (NGFF) WHERE the ever-increasing scale and complexity of image data and metadata would be contained for exchange31; blue bubble). B) The 4DN-BINA-OME specifications for WHAT hardware specifications, image acquisition settings, and quality control metrics should be reported articulate along three complexity axes: 1) Guideline Tiers: The three guideline Tiers are employed to scale reporting requirements with experimental complexity. 2) Model Core vs. Extensions: The use of the Core of the OME Data Model vs. one or more of the 4DN-BINA extensions allows capturing different microscopy modalities. 3) Metadata-requirement Levels: The distinction between Must use vs. Should use metadata fields, is used to define what information is needed for different reporting purposes (i.e., quality, reproducibility, sharing value). Depicted is the intersection between the three dimensions (OME Core + 4DN-BINA Basic and Calibration extensions ∩ Tier 2 ∩ All available fields) that would be appropriate to describe an experiment in which a wide-field microscope is used to capture the dynamics of viral particle trafficking within infected cells. ∩, intersection.
