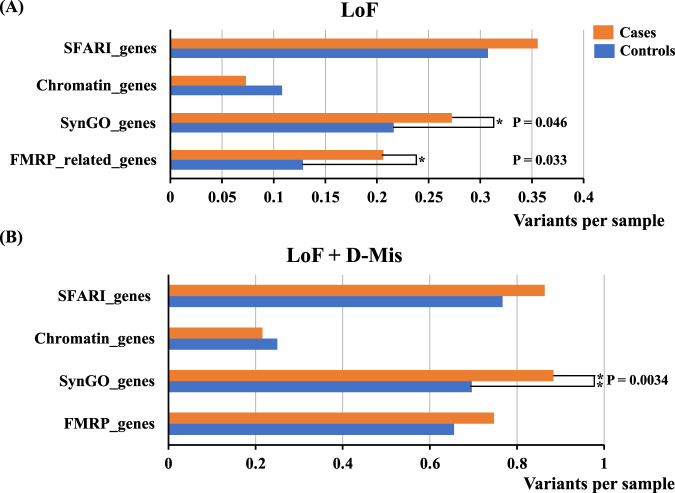
Fig. 2. Results of burden tests using gene sets related to ASD pathophysiology.
For the gene set burden tests, we used four gene sets: (1) FMRP target genes (FMRP_genes), (2) genes registered in the SynGO database (SynGO_genes), (3) genes encoding chromatin modifiers (Chromatin genes), (4) genes registered in the SFARI database (SFARI_genes). A The number of variants per sample for each gene set about LoF variants. B The number of variants per sample for each gene set about LoF + D-Mis variants. The LoF/LoF + D-Mis variants in the SynGO_genes were significantly enriched in ASD samples. The LoF variants in the FMRP_genes were significantly enriched in the ASD cases. The enrichment of the Chromatin_genes and SFARI_genes is not detected. An asterisk symbol indicates a nominal significant association (P-value < 0.05). Double asterisks indicate a nominal significant association (P-value < 0.01).