Figure 4. Glycan at Asn33 is present in all HA1 of group 1 except H9 and hinders 1D12 binding.
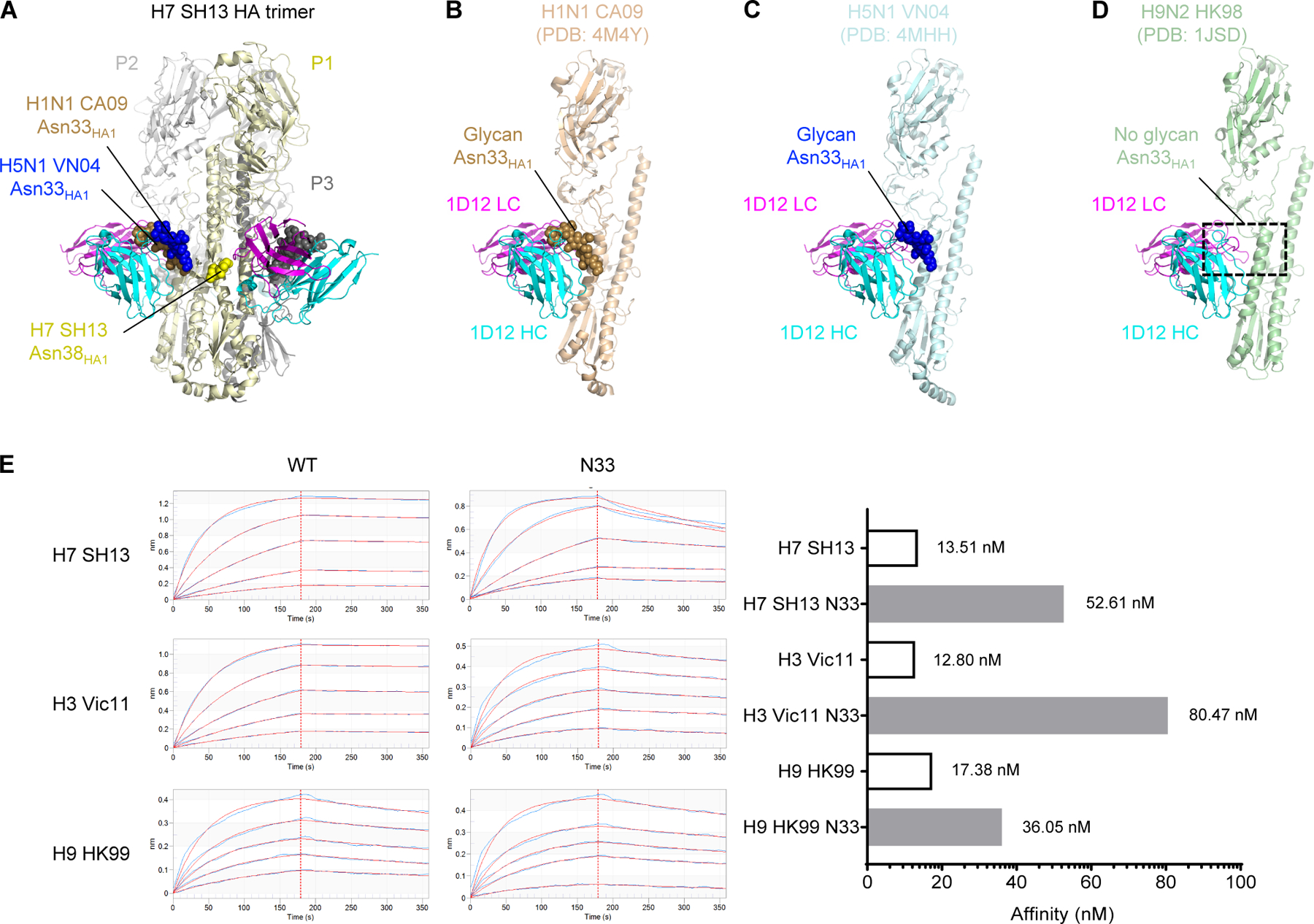
(A) A trimer of the H7 SH13 structure is shown with 1D12. The glycans at positions N33 and N38 of protomer 1 (P1, light yellow) are shown as spheres for this trimer and aligned trimers from H1N1 and H5N1. (B-D) Modelling of the 1D12 Fab onto the H1N1 CA09 HA (PDB: 4M4Y) (B), H5N1 VN04 HA (PDB: 4MHH) (C), and H9N2 HK98 (PDB: 1JSD) (D). The protein of protomer P2 is shown with the glycans from protomer P1. The presence of glycan at HA1 Asn33 in H1 CA09 and H5 VN04 resulted in clashes with the 1D12 Fab. Absence of glycan at HA1 Asn33 in H9 HK98 allowed 1D12 binding without clashes. (E) Octet affinity measurements of 1D12 Fab binding to wild-type group 2 (H3 and H7) as well as group 1 (H9) HAs with and without glycan Asn33 introduced. The KD values were shown on the right. Hemagglutinin analyte concentrations used 2-fold dilutions starting at 100 nM and ending with 6.25 nM.
