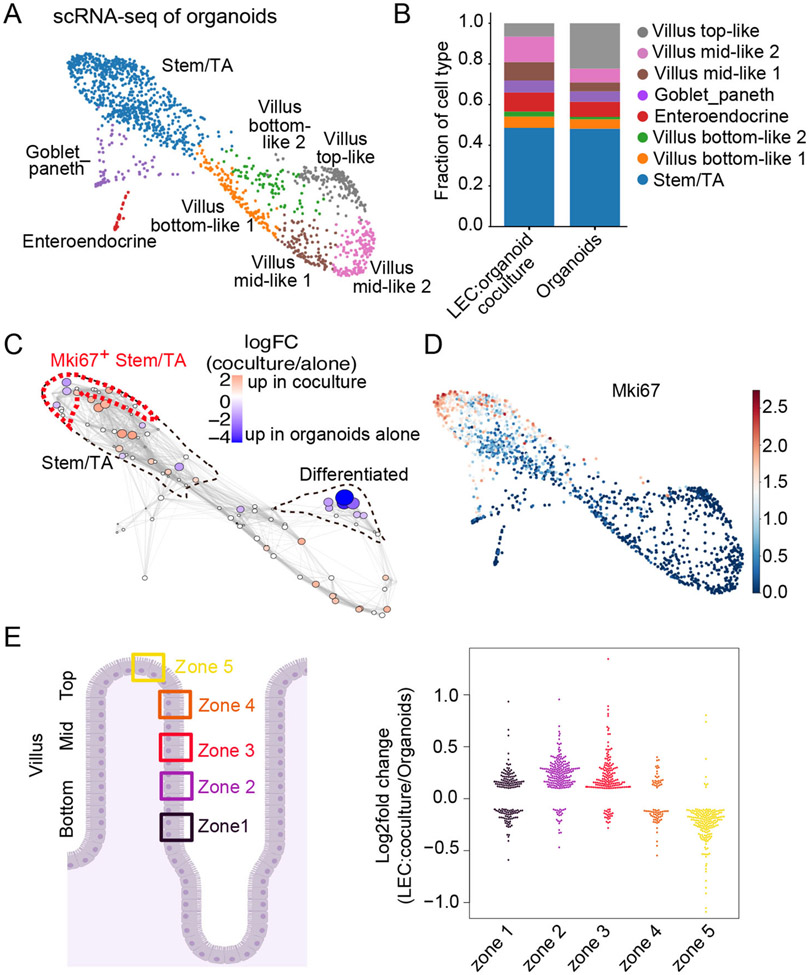
Figure 3: scRNA-sequencing supports a role for LECs in enhancing ISCs and reducing terminally differentiated enterocytes within organoids.
(A) Force-directed layout generated from scRNA-seq of organoids cultured with or without LECs. Cells are colored by cell type. (B) Bar plot shows the relative abundance of cell types per condition. (C) Differential abundance testing of cell states in scRNA-seq data using Milo (Dann et al., 2021). Nodes represent Milo neighborhoods, colored by their log2 fold change of cell abundance in LEC-cocultured organoids vs. organoids alone. Node size corresponds to the minus log10 false discovery rate (FDR). Stem/TA cells and differentiated enterocytes are outlined (dotted lines). Mki67+ population from (D) is outlined in red. (D) Force-directed layout colored by the log-transformed normalized gene expression values of Mki67 (cycling cells). (E) Log2 fold change of the expression of villus enterocyte zone genes (Moor et al., 2018) in cocultured and control organoids. Each dot represents a zone gene. Only significantly differentially expressed genes were visualized (absolute value of log2 fold change > 0.1 and FDR < 0.01).