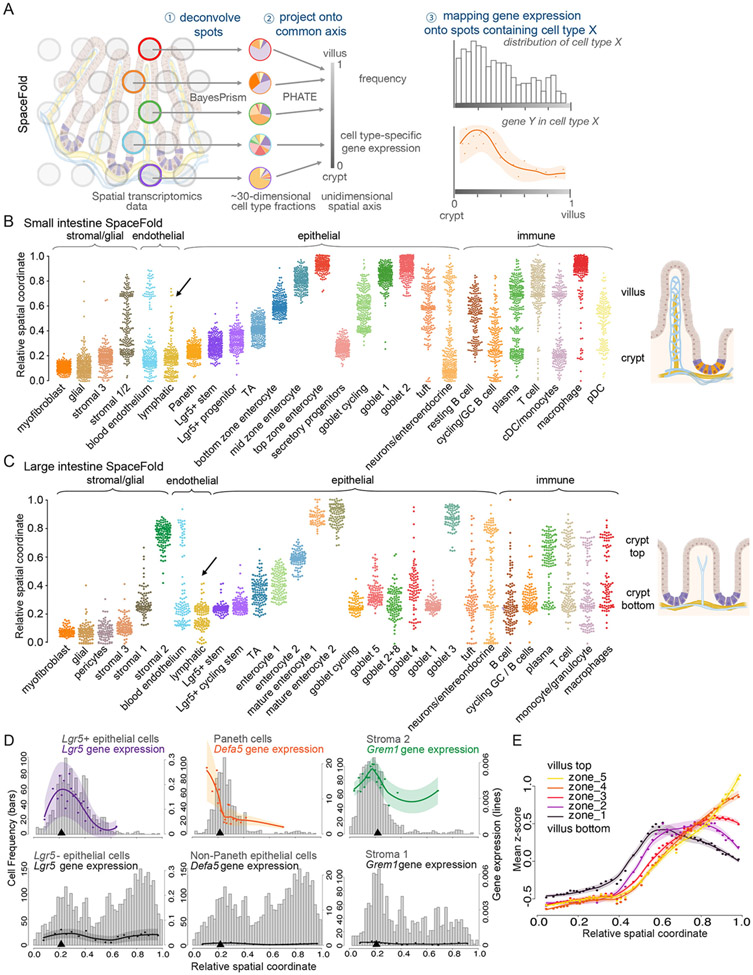
Figure 5. SpaceFold cartography reconstructs transcriptomes along crypt-villus and crypt axis.
(A) SpaceFold schematic. Each spatial spot was deconvolved using BayesPrism (step 1) to infer cell type fractions. Vectors of cell type fractions were reduced to a 1D projection that approximates its physical position along the crypt or crypt-villus axis (step 2). BayesPrism’s cell type-specific gene expression was used to generate trends along this axis for spots containing the cell type(s) of interest (step 3). (B-C) SpaceFold reveals the relative spatial coordinates of cell types, as identified from individual spots, along the SI crypt-villus (B) and LI crypt axis (C). Cell type annotation follows the cluster nomenclature in figures 4B-4C. Each dot represents a Visium spot containing the indicated cell type, plotted along its SpaceFold projection. Black arrow in B denotes computationally reconstructed lymphatic lacteals in the SI, absent in the LI in (C). (D) SpaceFold maps cell type-specific expression of known cell type markers onto the SI crypt-villus axis. X-axes mark the relative SpaceFold spatial coordinate. Histograms show the frequency of spots containing the selected cell types along the crypt-villus axis with corresponding Y-axes on the left. Lines mark smoothed mean values and shaded areas represent the mean ± 2 standard error of total (groups of cell types in left and center panels) or normalized (individual cell types in right panel) gene expression levels inferred by BayesPrism, corresponding to Y-axes on the right. Top panels show the predicted expression in groups of cell types or cell types expected to express the indicated marker genes, while bottom panels show the predicted expression in cell types not expected to express those genes. Black arrowheads indicate the spatial position of crypt-base lymphatics. (E) Intestinal villus epithelial cell zone gene expression distributed spatially along the projected SpaceFold crypt-villus axis. Similar to D, mean z-scores of the normalized expression of each group of zone markers in enterocytes are shown. Each dot represents the mean z-scores averaged over an interval of spatial spots binned by the SpaceFold coordinate. The x-coordinate of each dot represents the mean of the SpaceFold spatial coordinates in each bin. Lines mark the mean values fitted using local polynomial regression. Shaded areas represent the mean ± 2 standard error.