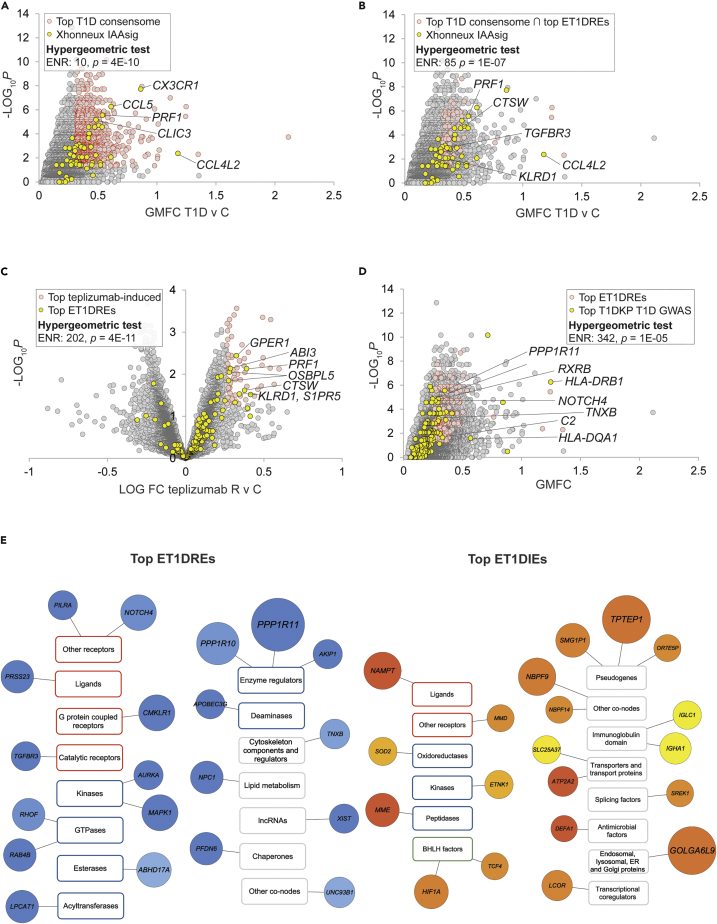
Figure 1.
Consensome feature level expression enrichment (C-FLEE) analysis identifies genes preferentially repressed or induced in T1D v control circulating immune cells
Selected transcripts at the intersection of the indicated gene sets are labeled. Refer to supplementary text for cut-offs used for hypergeometric tests.
(A) Scatterplot showing enrichment of Xhonneux IAAsig genes among the top-ranked genes in the T1D consensome.
(B) Scatterplot showing super-enrichment of Xhonneux IAAsig genes among the top-ranked ET1DREs in the T1D consensome.
(C) Volcano plot showing enrichment of top ET1DIE genes among top teplizumab responder (R) v control (C)-repressed genes in ABATE T1D cohort.
(E) Scatterplot showing enrichment of strong T1D GWAS candidates among top-ranked ET1DRE genes in the T1D consensome. (E) Selected top 20 ET1DRE (left panel) and ETD1IE (right panel) genes annotated using the three-tier (category, class, and family) SPP-controlled vocabulary {Ochsner, 2019 #83}. Circle size maps to enrichment p value: for reference, TPTEP is 1E-13 and DEFA1 is 4E-07. Light blue – dark blue (ET1DREs) and yellow-orange-red (ET1DIEs) color intensity maps to enrichment odds ratio: for reference in the ET1DREs, ABHD17A is 5.8 and PPP1R11 is 7.7; for reference in the ET1DIEs, IGLC is 7.3 and NAMPT is 11.8. Transcript class borders are color-coded according to their SPP classification category {Ochsner, 2019 #83} as follows: receptors (orange), enzymes (blue), transcription factors (green), and co-nodes (gray).