Abstract
PURPOSE
The Mayo-Baylor RIGHT 10K Study enabled preemptive, sequence-based pharmacogenomics (PGx)-driven drug prescribing practices in routine clinical care within a large cohort. We also generated the tools and resources necessary for clinical PGx implementation and identified challenges to be overcome. Furthermore, we measured the frequency of both common genetic variation for which clinical guidelines already exist and that of rare variation that could be detected by DNA sequencing, rather than genotyping.
METHODS
Targeted oligonucleotide-capture sequencing of 77 “pharmacogenes” was performed using DNA from 10,077 consented Mayo Clinic Biobank volunteers. The resulting predicted drug response related phenotypes for 13 genes, including CYP2D6 and HLA, affecting 21 drug-gene pairs, were deposited preemptively in the Mayo electronic health record (EHR).
RESULTS
For the 13 pharmacogenes of interest, the genomes of 79% of participants carried clinically actionable variants in 3 or more genes and DNA sequencing identified an average of 3.3 additional conservatively predicted deleterious variants that would not have been evident using genotyping.
CONCLUSIONS
Implementation of preemptive rather than reactive, sequence-based rather than genotype-based PGx prescribing revealed nearly universal patient applicability and required integrated institution-wide resources to fully realize individualized drug therapy and to demonstrate more efficient use of health care resources.
INTRODUCTION
Pharmacogenomics (PGx) is the study of genetically determined variation in individual response to drugs.1–3 PGx variants may impact either pharmacokinetics (PK), processes such as drug metabolism or transport that influence the concentration of a drug reaching its target, or pharmacodynamics (PD), variation in the target itself or processes downstream of the target.1,2 While many DNA sequences are known to influence drug response,1–3 PGx has not yet achieved broad clinical implementation. More recently, there have been multiple efforts to move toward that goal, from studies conducted by single institutions4 to those involving multiple institutions and multiple countries such as the “Ubiquitous Pharmacogenomics Consortium” that extends across the European Community5. There are numerous reasons for this slow pace of implementation including a requirement to educate providers, limited insurance reimbursement in the United States (U.S.), and a relative lack of prospective comparisons of preemptive test results versus reactive testing.
Numerous entities such as the Pharmacogenetics Knowledgebase (PharmGKB)6 and PharmVar7 have been established to serve as database repositories to collect PGx variants and provide underlying information including allele frequencies, metabolism pathways and the strength of associations between variants and clinical effects. Further resources including the Clinical Pharmacogenetic Implementation Consortium (CPIC),8 the Dutch Pharmacogenetic Working Group (DPWG)9 and others, are dedicated to the establishment of peer-reviewed clinical dosing guidelines based on diplotype-driven predictions of resulting drug response phenotypes. Various government entities such as the United States Food and Drug Administration (FDA)10 both publish dosing guidelines and exercise regulatory authority over testing and return of results. All of these databases catalog genomic variants that are relatively common, where there is sufficient data to support statistically robust conclusions as to their impact. However, over the past decade, it has been shown that rare variants are both greater in number and often demonstrate larger effect sizes than many common variants.11 A number of studies aimed at genes known to be involved in PGx also show that both common and rare variants are prevalent, influence clinical outcomes and that variants and allele frequencies vary across populations, suggesting that individual use of these data might have a broad and significant impact on drug prescribing practices worldwide.
A number of health care systems have begun to integrate “alerts” for PGx “drug-gene pairs” into their electronic health records (EHRs).12–14 Those alerts have sometimes been designed to inform the prescriber of the availability of PGx testing for the drug being prescribed, as is the case at the Mayo Clinic for a subset of drug-gene pairs (see Table 1 legend). However, this “reactive” approach requires that the prescriber order the PGx test and then wait for the result, a delay that could be avoided if genomic information for that patient had already been deposited in the EHR. In the ideal, a preemptive test would integrate PGx seamlessly into the clinical workflow, would only fire alerts for patients whose genomes carry the variant(s) of interest, would avoid delays in the initiation of drug therapy and would provide the prescriber with information for all sequence variants in the gene(s) of interest rather than merely a small number of commonly genotyped single nucleotide variants (SNVs) or structural variants such as indels (insertions or deletions) and CNVs (copy number variants).
Table 1.
Drug-Gene Pair Alerts implemented in the Mayo Clinic EHR by year of implementationa.
| Drug | Gene(s) | Year implemented |
|---|---|---|
| Abacavir | HLA-B*57:01 | 2013 |
| Azathiopurine | TPMT and NUDT15b | 2013 |
| Carbamazepine | HLA-B*15:02 and HLA-A*31:01c | 2013 |
| Codeine | CYP2D6 | 2013 |
| Mercaptopurine | TPMT and NUDT15b | 2013 |
| Tamoxifen | CYP2D6 | 2013 |
| Thioguanine | TPMT and NUDT15b | 2013 |
| Tramadol | CYP2D6 | 2013 |
| Allopurinol | HLA-B*58:01 | 2014 |
| Clopidogrel | CYP2C19 | 2014 |
| Simvastatin | SLCO1B1 | 2014 |
| Warfarin | CYP2C9 and VKORC1 | 2014 |
| Citalopram | CYP2C19 | 2015 |
| Escitalopram | 2015 | |
| Fluvoxamine | CYP2D6 | 2015 |
| Fluoxetine | CYP2D6 | 2015 |
| Paroxetine | CYP2D6 | 2015 |
| Venlafaxine | CYP2D6 | 2015 |
| Tacrolimus | CYP3A5 | 2016 |
| Capecitabine | DPYD | 2017 |
| Fluorouracil | DPYD | 2017 |
A subset of these alerts were designed to fire in a “reactive fashion”, i.e. recommending PGx testing in response to all initial prescriptions: TPMT and NUDT15 for thiopurines (mercaptopurine, azathioprine and thioguanine), HLA-B*57:01 for abacavir, HLA-B*15:02 and HLA-A*31:01 for carbamazepine in patients of Asian descent, HLA-B*58:01 for allopurinol in patients of Asian or African decent and CYP2D6 for tamoxifen. This was done to avoid physician “alert fatigue” that might have occurred if all of the alerts had been reactive. All other alerts currently fire only for patients who already have PGx information in the EHR. Between March 2015 and December 2018 these alerts fired a total of 6620 times. No comparable data are available after December 2018 because of Mayo Clinic’s implementation of a new EHR.
NUDT15 added in 2018 and assayed by genotyping.
HLA-A*31:01 added in 2018.
As a feasibility test and to identify challenges associated with the application of this approach to the clinical implementation of PGx guidance and—eventually—as a tool to study the clinical utility and economic benefit of preemptive sequence-based PGx, the Mayo Clinic and the Baylor College of Medicine Human Genome Sequencing Center (BCM-HGSC) collaborated to generate DNA sequence data for 77 known or candidate pharmacogenes using a capture panel and DNA from 10,077 patients who received their healthcare from the Mayo Clinic. Specifically, drug response phenotypes predicted for gene variants included in drug-gene pairs for which alerts currently fire at the Mayo Clinic were deposited in the EHR to determine whether preemptive sequence-based PGx testing might represent a step toward the broader incorporation of this aspect of clinical genomics into patient care. The DNA sequence information was generated under College of American Pathologists (CAP) and Clinical Laboratory Improvement Amendments (CLIA) standards. This effort was also designed to make it possible to stimulate PGx research studies by taking advantage of clinical drug response information present in the Mayo Clinic EHR. Table 1 lists the 21 drug-gene pairs for which PGx alerts fired across the Mayo Clinic system during the study as well as the year of their implementation. The 77 pharmacogenes that were sequenced for this study are listed in Table 2, with genes highlighted for which gene-related predicted phenotype status (e.g., “poor metabolizer”, etc.) were deposited preemptively in the EHR. We chose to clinically implement data for only those drug-gene pairs which had already undergone rigorous internal peer review for clinical utility—as described subsequently in the Methods, with the remaining genes being available for inclusion in future research studies to assess their potential clinical utility. The Mayo-Baylor RIGHT 10K Study also made it possible to identify significant institutional infrastructural and educational challenges associated with the implementation of this important aspect of clinical genomics.
Table 2.
The table lists the 77 pharmacogenes that were sequenced for the Mayo-Baylor RIGHT 10K Study. aThe “boxed and bold” genes are those for which predicted drug metabolism-drug response phenotype data were deposited in the Mayo Clinic EHR for RIGHT 10K subjects.
| ABCB1 | CYP2B6 | DPYD | GSTP1 | KCNH2 | RYR2 | SULT1A1 |
| ABCC4 | CYP2C8 | DRD2 | HLA-A (*31:01) | KIF6 | SCN1A | TP53 |
| ABCG2 | CYP2C9 | DRD3 | HLA-B (*15:02, *57:01, *58:01) | LDLR | SCN5A | TPMT |
| ADRB2 | CYP2C19 | DRD4 | HMGCR | LEP | SLC19A1 | TYMS |
| ANKK1 | CYP2D6 | EGFR | HNF1A | LEPR | SLC22A1 | UGT1A1 |
| CES1 | CYP2E1 | F5 | HNF4A | MTHFR | SLC22A2 | UGT1A3–10 exon1a |
| CFTR | CYP2J2 | FAAH | HTR2A | NAT2 | SLC6A4 | UGT2B15 |
| COMT | CYP3A4 | G6PD | HTR2C | OPRM1 | SLCO1B1 | UGT2B7 |
| CYP1A2 | CYP3A5 | GGCX | IFNL3 (IL28B) | PON1 | SLCO2B1 | VEGFA |
| CYP2A6 | CYP4F2 | GRIK4 | IGFBP7 | RYR1 | SOD2 | VKORC1 |
Please note that multiple UGT1 splice isoforms have been included and that results for CYP1A2, CYP3A4 and UGT1A1 were deposited to the EHR but are not yet part of drug-gene pair alerts at the Mayo Clinic.
MATERIALS AND METHODS
Study Participants
The 10,077 Mayo-Baylor RIGHT 10K study participants were volunteers who had donated biospecimens and health information to the Mayo Clinic Biobank for research purposes. Details regarding the design of the study have been described elsewhere.15 Demographic information for the study cohort is provided in Table 4.
Table 4:
Characteristics of the RIGHT 10K Participants
| Characteristic | n = 10,077 (Percentages) |
|---|---|
| Sex, n (%) | |
| Female | 6,146 (61.0) |
| Male | 3,931 (39.0) |
| Age on January 1, 2016, years, n (%) | |
| 18–24 | 58 (0.6) |
| 25–34 | 647 (6.4) |
| 35–44 | 824 (8.2) |
| 45–54 | 1,299 (12.9) |
| 55–64 | 2,067 (20.5) |
| 65–74 | 3,215 (31.9) |
| 75+ | 1,967 (19.5) |
| Race, n (%) | |
| White | 9,475 (94.0) |
| Non-White | 523 (5.2) |
| Black | 50 |
| Asian | 91 |
| AIAN | 16 |
| NHPI | 0 |
| Other and mixed | 366 |
| Unknown | 79 (0.8) |
| Ethnicity, n (%) | |
| Non-Hispanic | 9,959 (98.8) |
| Hispanic | 112 (1,1) |
| Unknown | 6 (0.1) |
| Self-reported education at time of Biobank Consent (2009–2017), n (%) | |
| High school graduate or GED or less | 1,261 (12.5) |
| Some college or Associates degree (including community college) | 2,935 (29.1) |
| Four year college graduate (Bachelor’s degree) | 1,991 (19.8) |
| Graduate or professional school | 3,845 (38.2) |
| Unknown | 45 (0.4) |
AIAN, American Indian or Alaska Native; NHPI, Native Hawaiian or Pacific Islander.
DNA Sequencing
Blood was collected and genomic DNA extracted at the Mayo Clinic for target-enriched capture sequencing at the BCM-HGSC Clinical Laboratory. A new PGx capture reagent (PGx-seq) was designed, constructed and validated to CAP-CLIA standards for clinical testing. This reagent targeted a combination of complete coding sequences for 77 pharmacogenes and variants present on both the Affymetrix DMET Plus (Affymetrix/Thermo Fisher Scientific, Santa Clara, CA) and Illumina VeraCode ADME (Illumina, San Diego, CA) array genotyping platforms not already addressed by the capture, together with supplementary known PGx and fingerprinting SNVs, as well as regional capture of the CYP2D6 locus that incorporated both of its nearby pseudogenes. In the development of both gene targets and software analysis, a previously characterized cohort of 512 samples was used to validate performance. Those analyses suggested that tag SNVs designed to identify the four HLA region allele-types of interest underperformed for one allele-type and, therefore, Omixon HLA Explore software (Omixon Biocomputing Ltd.; Budapest, Hungary) was tested and adopted for these loci. For CYP2D6, a software solution designed to identify structural and copy number variants was developed by the Mayo Clinic Personalized Genomics Laboratory (PGL) and implemented for these samples. The resulting average sample sequencing depth was greater than 490X. See Supplementary Methods for further details.
Drug-Gene Pair Alerts and Clinical Decision Support.
The Mayo Clinic Pharmacogenomics Task Force selected the drug-gene pair alert rules based on peer reviewed published guidelines from CPIC, DPWG, PharmGKB and the U.S. FDA as well as advice from intramural clinical specialists. This Task Force also developed the Clinical Decision Support (CDS) tools required to translate PGx assay results into EHR alerts.
Information Technology (IT)
The BCM-HGSC and the Mayo Clinic PGL collaborated to identify and “force call” (override variant caller software to return a locus-specific genotype regardless of conflict with the human reference) 310 potential variant sites defining currently known actionable alleles and developed translation lookup tables specifying drug response-related predicted phenotypes for all reportable genes except CYP2D6. For this gene, software developed in the Mayo PGL, CNVAR, was used to determine final CYP2D6 predicted phenotypes, or in rare cases, to refer samples for further testing and/or manual review. Scripts at Baylor were used to filter the Omixon software output to identify relevant HLA allele-types. Further scripts and modifications of existing data pipelines specific to the project were implemented at both institutions. See Supplementary Methods for further detail.
RESULTS
Introduction
The Mayo-Baylor RIGHT 10K Study was designed to test the long-term hypothesis that ready access at the point-of-care to prescribing recommendations based on a patient’s genetic composition would help to optimize that individual’s prescription drug therapy both short and long-term. We also sought to assess the rate of occurrence of rare variants within the PGx genes of participants and the added value of using DNA sequencing, rather than genotyping, for clinical testing. We anticipated that increases in drug efficacy and reduction in the rate of adverse events would result in better patient outcomes and enhanced health care economics as we accumulate cohort outcomes data moving forward. The clinicians caring for the 10,077 participants in the study, predominately primary care physicians, had not ordered PGx testing and, therefore, educational programs had to be designed for all healthcare team members as well as processes for the return of PGx results to both the healthcare team and to participating subjects.
Study Participant PGx DNA Variants
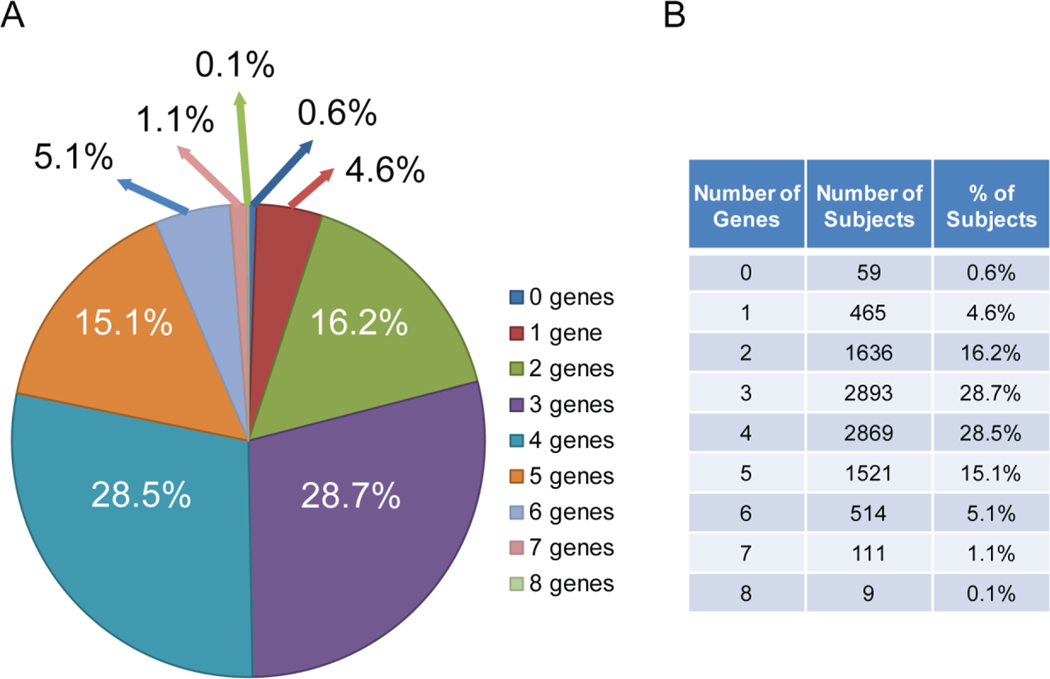
Targeted DNA sequencing for the 10,077 participants in the study, as anticipated, identified a large number of currently clinically actionable PGx variants as well as currently unclassified rare variants in the genomes of every patient (see Table 3)—both for all 77 genes and for the 13 genes for which alerts fired in the Mayo Clinic EHR during the study. The targeted genomic regions of all but 55 of the 10,077 participants included a clinically actionable variant in at least one of the 13 pharmacogenes included in the 21 drug-gene pair alerts (Table 1), while 79% had clinically actionable variants in three or more of those 13 genes, as depicted graphically in Figure 1. Therefore, all but 0.6% of the participants, depending on the drugs prescribed by their clinician, would potentially have benefitted from PGx information for just these 13 pharmacogenes.
Table 3.
The Total and Average Number of SNVs/Indels in the Sequenced Samples (n=10,077).
| All Calls | Deleterious Callsa: CADD/SIFTb/Intersection | ||||
|---|---|---|---|---|---|
| SNVs | Total Counts | Novel Counts | Total Counts | Novel Counts | |
| All 77 Genes | Number of SNVs | 10,205,546 | 9,997,316 | 462,915/360,310/190,311 | 426,384/346,108/176,148 |
| SNVs/Sample | 1,012.8 | 992.1 | 45.9/35.8/18.9 | 42.3/34.4/17.5 | |
| 13 Alert Genes | Number of SNVs | 1,388,855 | 1,219,760 | 93,875/92,352/44,614 | 73,987/80,835/33,136 |
| SNVs/Sample | 137.8 | 121.0 | 9.3/9.2/4.4 | 7.3/8.0/3.3 | |
| INDELS | Total Counts | Novel Counts | Total Counts | Novel Counts | |
| All 77 Genes | Number of Indels | 620,215 | 576,068 | 11,749/0/0 | 1,697/0/0 |
| Indels/Sample | 61.6 | 57.2 | 1.2/0/0 | 0.2/0/0 | |
| 13 Alert Genes | Number of Indels | 69,787 | 56,346 | 5,509/0/0 | 0/0/0 |
| Indels/Sample | 6.9 | 5.6 | 0.6/0/0 | 0/0/0 | |
Novel Counts are total SNV/Indel variant counts excluding those included in 133 CPIC actionable variants included in the Mayo Drug-Gene Pair Alerts plus stop gain, stop loss and frameshift variants. For this analysis, “deleterious” has been defined as having a CADD score > 2017 and/or a SIFT score < 0.05.18
SIFT output is limited to frameshift/non-frameshift for indels.
Figure 1.
The Figure shows the number of genes that contained clinically actionable genomic variants for the 13 genes included in the Drug-Gene Pair Alerts listed in Table 1 that were observed in each of the 10,077 RIGHT 10K Study subjects as well as the percentage of study subjects included in each group. A. The “pie chart” shows these data graphically while the table in B lists the information upon which the pie chart is based.
Sequencing vs. Genotyping
We chose a targeted DNA sequence-based assay for this project to ensure that we tested all target genes comprehensively. The alternative DNA genotyping arrays currently available are, of necessity, designed to only detect variants that have been previously identified. Consequently, available arrays generally represent relatively more common variation and, conversely, fail to represent less common genomic variants which could—in aggregate—have significant clinical relevance.16
As a proxy for a comparison with the collection of variants available on arrays, we filtered our DNA sequence results to remove common variants already categorized by CPIC as clinically actionable (Table 3). After filtering, on average, each participant carried 127 additional SNV and Indel variants for the 13 genes listed in Table 1. Computer analysis (e.g., CADD: Combined Annotation Dependent Depletion and SIFT: Sorting Intolerant From Tolerant) scores17,18 identified an average of 7.3 and 8.0 CADD and SIFT variants respectively and 3.3 variants when intersected, in each participant as “probably deleterious”, suggesting a potential clinical impact. A recent report of functional testing by “deep mutational scanning” in a separate cohort combined with functional validation of the results, found that 19 of 109 CYP2C9 and 36 of 121 CYP2C19 variants that had been identified by sequencing the genomes of large populations, displayed “severely damaging” phenotypes with protein expression that was less than 25% of that present for “wild type” alleles.19 CYP2C9 and CYP2C19 are pharmacogenes that play important roles in variable clinical response to drugs that include warfarin, clopidogrel and a number of psychiatric drugs. Six of the subjects included in the present study carried functionally severely damaging variants in CYP2C9 other than those that are “usually genotyped”, while the genome of one subject carried two such variants. The genomes of twenty-five of our participants included one functionally severely damaging variant in their CYP2C19 gene beyond those usually genotyped, while one subject carried two such variants. All of these cases would have been missed if only “standard genotyping” had been applied. Variants and alleles with unknown function were evaluated using a modification of ACMG variant interpretation criteria. Predicted phenotypes for variants of uncertain significance (VUS) were expressed as a range (See Supplementary Methods).20
Application to Current Therapy
As a first step toward implementation, clinicians caring for patients already prescribed medications influenced by variants in genes included in the Mayo Clinic drug-gene pair alerts were informed whenever these initial results suggested that a patient’s drug therapy could potentially be improved by dose adjustment or alternative therapy. If Mayo Clinic pharmacists concluded that the PGx test results indicated either a “semi-urgent” (i.e., the drug had the potential to cause serious harm) or a “clinically actionable” (i.e., the drug had the potential to cause an adverse reaction or significantly altered efficacy) need to inform the prescriber, eConsults were sent to the primary care provider. Semi-urgent eConsults were sent for 61 patients. The drugs involved were clopidogrel for 41 of the 61 patients (67%), citalopram 9/61 (15%), escitalopram 7/61 (11%), tramadol 2/61 (3%), fluorouracil 1/61 (2%), and allopurinol 1/61 (2%). Providers for those patients accepted 54% of the pharmacists’ semi-urgent eConsult recommendations. Viewed more globally, a total of 2782 clinically actionable eConsults were sent out to providers based on the 10K sequencing data—a figure that begins to provide insight into the potential benefit if PGx information had been available at the time that medications for those patients had initially been prescribed. Obviously, most of these patients had been on their current therapy regimen for some time, so their drug therapy might have already been altered in response to either the occurrence of an adverse reaction or lack of efficacy. In addition, the Mayo Clinic has begun collecting evidence of improvement in outcomes traced to the application of the RIGHT 10K results to participant clinical care. Among several examples in psychiatric patients, one participant, found to be a CYP2C19 ultrarapid metabolizer, was switched from an ineffective combination of escitalopram supplemented with bupropion to bupropion monotherapy and is now reported to be in full remission from major depressive disorder. In another example, a participant on combination therapy was found to be a CYP2D6 poor to intermediate metabolizer, resulting in the recommendation that tramadol be replaced or eliminated, resulting in the alleviation of associated dizziness.
Pharmacists and PGx Implementation
We found that a team-based approach entailing the involvement of PGx-trained pharmacists, information technology (IT) support for the development of decision support rules and alerts, and effective PGx education programs were all required for the success of this implementation effort. Specifically, the development of PGx test reports that were easily understood by clinicians was essential, as well as ensuring that clinicians had access to those reports prior to display in patient portals. The Mayo Clinic’s Department of Pharmacy played a key role in many of these processes, but initial expertise in PGx among pharmacists was highly variable. Therefore, a “train-the-trainer” model21 was applied to ensure that the majority of pharmacists across the Mayo Clinic had been trained in PGx, with early adopters serving as trainers for their colleagues. The Department also established an electronic consultation and recommendation process (the eConsults referred to above) to provide patient-specific PGx guidance to providers. Those efforts resulted in a total of 392 of Mayo’s 452 Minnesota licensed pharmacists being trained in PGx, the fact that the Mayo Rochester Pharmacy now includes three full-time-PGx specialists, the establishment of an annual Pharmacogenomics Workshop that rotates among the three major Mayo Clinic campuses in Minnesota, Florida and Arizona, a post-graduate year two residency (PGY-2) training program in Pharmacogenomics for Doctor of Pharmacy (PharmD) graduates and the launch of an online PGx Certificate Program.
PGx Education for Medical Staff
Previous studies have reported that clinicians often report that “lack of education” is a major factor limiting their ability to use PGx clinically.22–25 To help address this challenge, multidisciplinary PGx educational content was developed for both practitioners26 and pharmacists as outlined above. Furthermore, critical components of this content were incorporated into AskMayoExpert (AME), an institutional online knowledge resource that provides Mayo clinicians with point-of-care information on a wide variety of clinical topics. A direct link to the appropriate AME PGx topic was integrated into each EHR drug-gene pair alert together with information on both alternative medications and reference to Mayo Clinic experts who could provide assistance. Finally, case-based education for pharmacists, nurses and other providers was also developed.
RIGHT 10K Study and PGx Research.
The RIGHT 10K Study also provided a broad foundation for PGx discovery. The availability of DNA sequence information for 77 pharmacogenes joined with clinical data in the EHR created an unusual opportunity for PGx research. The sequence data were made available to Mayo Clinic investigators based on the submission of a short research protocol that underwent IRB and scientific peer review. We have approved 30 protocols that address PGx across a broad spectrum of drug therapies (see Supplementary Table 1). An unforeseen bonus of this approach to access has been the fact that the investigators leading those 30 studies have often become both experts in and advocates for PGx within their individual clinical departments and divisions—a development that has assisted with the clinical acceptance and use of PGx at the Mayo Clinic.
PGx 10K Participant Survey
A survey designed to query knowledge of and attitudes toward PGx was sent to half of the RIGHT 10K Study participants, and 4629 (92.8%) of the invited participants returned the survey. The survey was sent to participants after they had been consented but before they had received their PGx results. Respondents felt that PGx results would help them avoid exposure to medications that might be harmful (Quite Valuable to Extremely Valuable, 94.6%), but relatively few (<18%) expected that their prescription medications or dosages would have to be changed based on the PGx results. In addition, respondents reported low levels of concern regarding potential disruption of their ongoing care and 76% had no or few concerns about the ability of their physician to integrate those results into their care. A follow-up survey is planned after all participants have had an opportunity to discuss the PGx results with their care givers.
DISCUSSION
PGx has long represented one of the most compelling avenues toward bringing DNA-based individualized medicine to clinical care.1–3 Variability in response to medication has long been observed and expected among both providers and patients and genetic variability underlying this phenomenon is common. While a small number of mainly academic medical centers have adopted limited PGx testing protocols and although momentum is slowing building within a number of provider networks, broad implementation remains slow for several reasons. These include lack of overall knowledge of the topic and, therefore, acceptance among medical providers at all levels, challenges in translating test results to dosing recommendations in patients’ EHRs while not interrupting the clinical workflow and, in the U.S. in particular, a reluctance to date by both private insurance and government entities to reimburse the relatively minor cost involved in testing. The Mayo-Baylor RIGHT 10K Study represents a systematic effort to integrate preemptive DNA sequence-based PGx panel testing into clinical workflows across a large medical center incorporating a large cohort of patients. This study both builds on previous work and addresses challenges not generally taken on by others. As demonstrated by previous analyses of both the 1000 Genomes Project27 and more recently, the UK Biobank28 and anticipated here, we found that nearly all of the 10,077 study participants could potentially have benefitted from preemptive PGx data and its attendant interpretation deposited in the EHR—depending on the drugs that their clinician might prescribe. Of importance, we chose to apply targeted DNA sequencing rather than genotyping to allow us to capture less common genomic variants which could by themselves, or in combination, have significant clinical relevance and did so at a cost that barely exceeded the cost of genotyping.
We found that the average patient harbored at least three current clinically actionable variants and a conservatively estimated three further variants deemed probably deleterious for only the thirteen genes included in our study. The utility of the sequence data was further confirmed by the observation that 55 of 230 variants that we observed in the sequence data for CYP2C9 and CYP2C19 were recently classified as functionally significant19 and are unlikely to be assayed using currently available genotyping arrays. While the remaining variants are currently classified as VUS, ongoing functional testing is likely to provide reclassification for some of those variants in the future. Clinical testing by sequencing, rather than via genotyping arrays ensures that as a variant is reclassified, individuals can be informed of those results without the need for repeat testing with new reagents. We acknowledge a geographically driven bias in our cohort as reflected in the Northern European inheritance present for the vast majority of our samples (94%). As a result, we would anticipate that our data for uncharacterized variant frequency represents a lower boundary when these tools are applied to more diverse populations, as outlined by others.28
Our study developed both reagents and data analysis software tools that allowed the successful assay and reporting of predicted phenotypes across two very important but highly polymorphic and difficult assay targets, CYP2D6 and the human HLA region. The former is known to be involved in the metabolism or processing of up to 25% of all drugs currently marketed and the latter is known to be involved in several of the most serious adverse event episodes observed to date. There is little doubt that these loci will continue to play major roles in PGx going forward.
The RIGHT 10K Study also served to highlight a series of challenges associated with the clinical implementation of PGx. For example, we revealed the need to identify health care team interpreters, a role played in this study by Mayo Clinic pharmacists, to act as a conduit between the data, physicians and patients. Additionally, there was a critical need for IT support to build and implement the data pipelines that enable the identification of multiple DNA variant types and the translation of diplotypes to alleles and clinical decision support software that integrates smoothly with at least one and perhaps several EHR packages. We also learned where needs existed and developed solutions within Mayo’s educational infrastructure. Approaches to address these challenges will differ depending on the local environment of individual medical institutions or provider networks, but having professional staff who can play the interpretive, evaluative and educational role taken on by the Mayo Clinic pharmacists, such as physicians assistants, nurse practitioners or genetic counselors in other healthcare institutional settings, plus the availability of clinical decision support tools will be required to deliver PGx information to clinical staff quickly, clearly and without requiring that they be familiar with genomic science or the underlying data.
In summary, the Mayo-Baylor RIGHT 10K Study strongly suggests that preemptive sequence-based PGx panel implementation can be useful clinically—especially if delivered at the point-of-care when drugs are being prescribed--and that it would apply to almost every patient. These results may also be helpful to other medical centers as they consider how or whether they wish to implement preemptive sequence-based PGx panels. This study also serves as an important stepping-stone in the development of the infrastructure necessary to fully utilize the coming incorporation of whole genome sequencing in helping to guide clinical care. We should point out that the United States FDA has voiced some concerns with regard to PGx, particularly direct to consumer PGx testing and the possible extension of the application of test results beyond supporting clinical evidence.16,29 All testing described here was conducted in a CAP-CLIA environment with clinician supervision of every aspect of the study. Furthermore, all test interpretations rested on validated consensus-based guidelines.6,8–10 It should also be emphasized that PGx will continue to evolve. For example, the future will almost certainly include the application of machine learning-based predictive algorithms that utilize both clinical and genomic information joined with other types of data that will extend well beyond genotypes for the small number of genes included in the drug-gene pair alerts listed in Table 1.30 Finally, although PGx has not yet been broadly adopted in the clinic, it clearly offers the promise of ultimately becoming a standard component of clinical practice that will help the healthcare team optimize pharmacotherapy for all patients.
Supplementary Material
ACKNOWLEDGEMENTS
Funding/Support:
This work was supported by the Mayo Clinic Center for Individualized Medicine, the Mayo Clinic Robert D. and Patricia E. Kern Center for the Science of Health Care Delivery, Mayo Clinic Pharmacy Services and by the Augeo Foundation, the Satter Foundation, Bernard E. and Edith B. Waterman, William G. Little, and Roche Diagnostics Corporation, NSF 2041339 and by National Institutes of Health grants U19 GM61388 (The Pharmacogenomics Research Network), R01 GM28157, U01 HG005137, R01 GM125633, R01 AG034676 (The Rochester Epidemiology Project), and U01 HG06379 and U01 HG06379 Supplements [The Electronic Medical Record and Genomics (eMERGE) Network].
Role of the Funder/Sponsor:
The funders/sponsors had no role in the design and conduct of the study; collection, management, analysis and interpretation of the data; preparation, review or approval of the manuscript; or the decision to submit the manuscript for publication.
ETHICS DECLARATION
Conflict of Interest Disclosures:
Drs. Liewei Wang, Black and Weinshilboum are co-founders of and stockholders in OneOme, LLC, used only to return results to study participants.
Footnotes
AUTHOR INFORMATION
Conceptualization: Liewei.W., S.J.B., R.M.W., R.A.G, E.B., S.E.S; Data Curation: E.V., X.Q., J.M.V., C.M.F., A.S.; Formal Analysis: Liewei.W., S.J.B., D.M.M., S.E.S., X.Q., K.W., T.-J.W., Q.W., D.K., J.L.BIII., A.M.M., M.B.K., K.J.H., H.S., S.E.P., J.E.O., K.R.K., N.B.L., H.L., Liwei.W., G.S.L., B.J.B., R.R.F., Y.Z., D.J.J.,M.A.H., S.M.A., M.E.M., R.J.; Funding Acquisition: R.M.W., R.A.G.; Investigation: S.J.B., R.M.W., D.M.M., S.E.S., J.L.BIII., A.M.M., T.B.C.; Methodology: Liewei.W., S.J.B., M.W., J.L.S., R.M.W., E.B., D.M.M., S.E.S., E.V., X.Q., J.H., C.L.K., K.W., J.L.BIII., A.M.M., A.E.B., B.E.M., J.M.S., M.L.K., K.E.K., K.K., W.T.N., J.G., H.M., P.J.C., A.K., A.E., D.B., M.J.S., C.G., S.E., A.U., Leah.W., S.C., E.A.N., S.L., M.E., S.G.P., N.P., M.F., F.S., M.G., I.K., K.L., R.W., J.A.A., E.T.M., J.A.W., C.M.F., R.M.E., J.D.Z., J.R.H., C.A.K., R.W.H., J.L.T., T.B.C., T.M.M., C.R.R.V., S.R.S., I.A., M.E.B., S.R.F., A.G., P.L.G., T.M.J., P.T., M.B., K.F.-S., N.R.N., A.K.R., D.M.R., J.W., R.R.S.; Project Administration: Liewei.W., S.J.B., L.A.J, M.W., T.S., J.L.S., R.M.W., R.A.G., E.B., D.M.M., S.E.S., C.L.K., V.K., J.L.BIII., A.M.M., W.T.N., J.G., P.J.C., S.E., K.L., L.J.O., E.T.M., J.A.W., T.B.C., T.M.M., C.R.R.V., R.R.S.; Resources: S.J.B., H.D., R.R., S.D., W.L., G.C., L.Z., A.E.B., J.M.V, S.C.Z., S.B., C.G., M.L.P., J.E.P., J.E.O., E.J.S., A.T.B., C.H.; Software: M.W., E.V., X.Q., K.W., T.-J.W., J.L.BIII., K.J.H., H.S., S.E.P., J.M.V., J.S., A.S., T.H.K., J.L.R., M.H., K.B., B.R., I.S.; Supervision: Liewei.W., S.J.B., L.A.J., M.W., T.S., J.L.S., R.M.W., R.A.G., E.B., D.M.M., S.E.S., E.V., J.H., K.W., H.D., R.R., W.L., J.L.BIII., A.E.B., K.K., W.T.N., H.M., P.J.C., M.E., S.G.P., N.P., M.F., F.S., M.G., I.K., K.L., R.W., L.J.O., T.B.C., T.M.M., C.R.R.V., I.A., M.E.B., R.R.S., N.B.L.; Validation: D.M.M., S.E.S., E.V., X.Q., J.H., C.L.K., V.K., K.W., T.-J.W., R.R., S.D., W.L., G.C., L.Z., Q.W., D.K., J.L.BIII., M.B.K.; Writing-original draft: Liewei.W., S.J.B., R.M.W., R.A.G., S.E.S., E.V., X.Q., J.H., V.K.; Writing – review & editing: All Authors.
ETHICS STATEMENT
All participants in this study were recruited from the Mayo Clinic Biobank and informed consent was obtained under Mayo Clinic Institutional Review Board approval. Participants agreed to the use of their stored Biobank samples for clinical pharmacogenomic testing, deposit of pharmacogenomic results into their EHR for clinical use and use of deidentified pharmacogenomic data for research. For more information on both the Biobank (https://www.mayo.edu/research/centers-programs/mayo-clinic-biobank/about/governance-oversight) and the RIGHT cohort, please see Bielinski, et al.15
ClinicalTrial.gov identifier: NCT03803293.
Publisher's Disclaimer: This is a PDF file of an article that has undergone enhancements after acceptance, such as the addition of a cover page and metadata, and formatting for readability, but it is not yet the definitive version of record. This version will undergo additional copyediting, typesetting and review before it is published in its final form, but we are providing this version to give early visibility of the article. Please note that, during the production process, errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DATA AVAILABILITY
As outlined by Bielinski, et al.15, the RIGHT cohort is a resource for pharmacogenomic research31. As stated above, a RIGHT Data Access Committee has been created to review data requests for use of RIGHT data. External access to the data is facilitated by the Mayo Clinic Biobank (https://www.mayo.edu/research/centers-programs/mayo-clinic-biobank/ overview). All potentially damaging variants observed in this study are presented with annotation in Supplementary Table 2. The software developed in this study primarily represents output/input wrappers or filtering scripts and is described in Supplementary Methods. The privately developed CNVAR software (submitted for patent) was conceived and refined based on the genomic targets specified by the PGx-seq reagent and would need extensive adjustments for other captures. However, those interested in using CNVAR may contact the authors to determine whether the software could be applied to their data set. Omixon HLA Explore™ is commercially available, while the remaining underlying software is available at https://www.hgsc.bcm.edu/software.
REFERENCES
- 1.Weinshilboum RM, Wang L. Pharmacogenomics: Precision Medicine and Drug Response. Mayo Clin Proc. 2017;92(11):1711–1722. doi: 10.1016/j.mayocp.2017.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang L, McLeod HL, Weinshilboum RM. Genomics and drug response. N Engl J Med. 2011;364(12):1144–1153. doi: 10.1056/NEJMra1010600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roden DM, Van Driest SL, Wells QS, Mosley JD, Denny JC, Peterson JF. Opportunities and Challenges in Cardiovascular Pharmacogenomics: From Discovery to Implementation. Circ Res. 2018;122(9):1176–1190. doi: 10.1161/CIRCRESAHA.117.310965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marrero RJ, Cicali EJ, Arwood MJ, et al. How to Transition from Single-Gene Pharmacogenetic Testing to Preemptive Panel-Based Testing: A Tutorial. Clin Pharmacol Ther. 2020;108(3):557–565. doi: 10.1002/cpt.1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Wouden CH, Cambon-Thomsen A, Cecchin E, et al. Implementing Pharmacogenomics in Europe: Design and Implementation Strategy of the Ubiquitous Pharmacogenomics Consortium. Clin Pharmacol Ther. 2017;101(3):341–358. doi: 10.1002/cpt.602 [DOI] [PubMed] [Google Scholar]
- 6.Barbarino JM, Whirl-Carrillo M, Altman RB, Klein TE. PharmGKB: A worldwide resource for pharmacogenomic information. Wiley Interdiscip Rev Syst Biol Med. 2018;10(4):e1417. doi: 10.1002/wsbm.1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaedigk A, Ingelman-Sundberg M, Miller NA, Leeder JS, Whirl-Carrillo M, Klein TE, PharmVar Steering Committee. The Pharmacogene Variation (PharmVar) Consortium: Incorporation of the Human Cytochrome P450 (CYP) Allele Nomenclature Database. Clin Pharmacol Ther. 2018;103(3): 399–401. doi: 10.1002/cpt.910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Relling MV, Klein TE, Gammal RS, Whirl-Carrillo M, Hoffman JM, Caudle KE. The Clinical Pharmacogenetics Implementation Consortium: 10 Years Later. Clin Pharmacol Ther. 2020; 107(1):171–175. doi: 10.1002/cpt.1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swen JJ, Nijenhuis M, van Rhenen M, et al. Pharmacogenetic Information in Clinical Guidelines: The European Perspective. Clin Pharmacol Ther. 2018;103(5):795–801. doi: 10.1002/cpt.1049 [DOI] [PubMed] [Google Scholar]
- 10.FDA. Table of Pharmacogenomic Biomarkers in Drug Labeling. 08/18/2020. https://www.fda.gov/drugs/science-and-research-drugs/table-pharmacogenomic-biomarkers-drug-labeling (accessed 29 January 2020).
- 11.Lupski JR, Belmont JW, Boerwinkle E, Gibbs RA. Clan genomics and the complex architecture of human disease. Cell. 2011; 147(1):32–43. doi: 10.1016/j.cell.2011.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Donnell PH, Wadhwa N, Danahey K, et al. Pharmacogenomics-Based Point-of-Care Clinical Decision Support Significantly Alters Drug Prescribing. Clin Pharmacol Ther. 2017;102(5): 859–869. doi: 10.1002/cpt.709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hicks JK, Stowe D, Willner MA, et al. Implementation of Clinical Pharmacogenomics within a Large Health System: From Electronic Health Record Decision Support to Consultation Services. Pharmacotherapy. 2016;36(8):940–948. doi: 10.1002/phar.1786 [DOI] [PubMed] [Google Scholar]
- 14.Caraballo PJ, Bielinski SJ, St Sauver JL, Weinshilboum RM. Electronic Medical Record-Integrated Pharmacogenomics and Related Clinical Decision Support Concepts. Clin Pharmacol Ther. 2017;102(2):254–264. doi: 10.1002/cpt.707 [DOI] [PubMed] [Google Scholar]
- 15.Bielinski SJ, St Sauver JL, Olson JE, et al. Cohort Profile: The Right Drug, Right Dose, Right Time: Using Genomic Data to Individualize Treatment Protocol (RIGHT Protocol). Int J Epidemiol. 2020;49(1):23–24k. doi: 10.1093/ije/dyz123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.CPIC. FDA and Pharmacogenomics. 8/02/2019 2019. https://cpicpgx.org/wp-content/uploads/2019/08/fda-and-pgen-2019.pdf (accessed 27 April 2020).
- 17.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. doi: 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–81. doi: 10.1038/nprot.2009.86 [DOI] [PubMed] [Google Scholar]
- 19.Zhang L, Sarangi V, Moon I, et al. CYP2C9 and CYP2C19: Deep Mutational Scanning and Functional Characterization of Genomic Missense Variants. Clin Transl Sci. 2020;13(4):727–742. doi: 10.1111/cts.12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopes JL, Harris K, Karow MB, et al. Targeted Genotyping in Clinical Pharmacogenomics: What is Missing? J Mol Diagn. 2021; accepted for publication [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Formea CM, Nicholson WT, Vitek CR, et al. Implementation of a pharmacogenomics education program for pharmacists. Am J Health Syst Pharm. 2018;75(23):1939–1946. doi: 10.2146/ajhp170771 [DOI] [PubMed] [Google Scholar]
- 22.Stanek EJ, Sanders CL, Taber KA, et al. Adoption of pharmacogenomic testing by US physicians: results of a nationwide survey. Clin Pharmacol Ther. 2012;91(3):450–458. doi: 10.1038/clpt.2011.306 [DOI] [PubMed] [Google Scholar]
- 23.St Sauver JL, Bielinski SJ, Olson JE, et al. Integrating Pharmacogenomics into Clinical Practice: Promise vs Reality. Am J Med. 2016;129(10):1093–1099.e1. doi: 10.1016/j.amjmed.2016.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johansen Taber KA, Dickinson BD. Pharmacogenomic knowledge gaps and educational resource needs among physicians in selected specialties. Pharmgenomics Pers Med. 2014;7: 145–162. doi: 10.2147/PGPM.S63715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haga SB, Burke W, Ginsburg GS, Mills R, Agans R. Primary care physicians’ knowledge of and experience with pharmacogenetic testing. Clin Genet. 2012;82(4):388–394. doi: 10.1111/j.1399-0004.2012.01908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giri J, Curry TB, Formea CM, Nicholson WT, Rohrer Vitek CR. Education and Knowledge in Pharmacogenomics: Still a Challenge? Clin Pharmacol Ther. 2018;103(5):752–755. doi: 10.1002/cpt.1019 [DOI] [PubMed] [Google Scholar]
- 27.Wright GEB, Carleton B, Hayden MR, Ross CJD. The global spectrum of protein-coding pharmacogenomic diversity. Pharmacogenomics J. 2018;18(1):187–195. doi: 10.1038/tpj.2016.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McInnes G, Lavertu A, Sangkuhl K, Klein TE, Whirl-Carrillo M, Altman RB. Pharmacogenetics at Scale: An Analysis of the UK Biobank. Clin Pharmacol Ther. 2020; Epub ahead of print. doi: 10.1002/cpt.2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DEN180028 FDA. Evaluation of Automatic Class III Designation for “The 23andMe Personal Genome Service (PGS) Pharmacogenomic Reports. https://www.accessdata.fda.gov/cdrh_docs/reviews/DEN180028.pdf (accessed 27 April 2020).
- 30.Athreya AP, Neavin D, Carrillo-Roa T, et al. Pharmacogenomics-Driven Prediction of Antidepressant Treatment Outcomes: A Machine-Learning Approach With Multi-trial Replication. Clin Pharmacol Ther. 2019;106(4):855–865. doi: 10.1002/cpt.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bielinski SJ, Olson JE, Pathak J, et al. Preemptive genotyping for personalized medicine: design of the Right Drug, Right Dose, Right Time-using Genomic Data to Individualize Treatment Protocol. Mayo Clin Proc. 2014;89:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
As outlined by Bielinski, et al.15, the RIGHT cohort is a resource for pharmacogenomic research31. As stated above, a RIGHT Data Access Committee has been created to review data requests for use of RIGHT data. External access to the data is facilitated by the Mayo Clinic Biobank (https://www.mayo.edu/research/centers-programs/mayo-clinic-biobank/ overview). All potentially damaging variants observed in this study are presented with annotation in Supplementary Table 2. The software developed in this study primarily represents output/input wrappers or filtering scripts and is described in Supplementary Methods. The privately developed CNVAR software (submitted for patent) was conceived and refined based on the genomic targets specified by the PGx-seq reagent and would need extensive adjustments for other captures. However, those interested in using CNVAR may contact the authors to determine whether the software could be applied to their data set. Omixon HLA Explore™ is commercially available, while the remaining underlying software is available at https://www.hgsc.bcm.edu/software.