Figure 3.
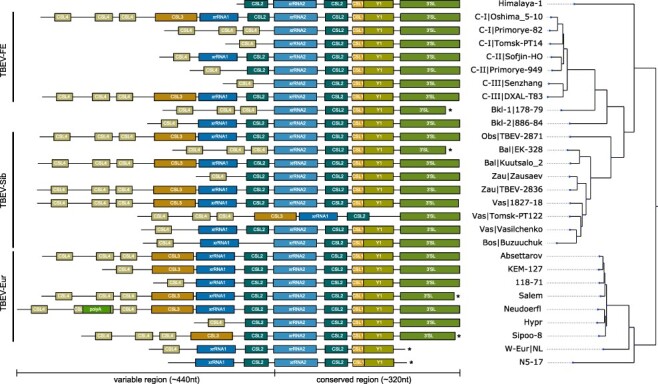
Annotated 3ʹUTRs of representative TBEV strains. Strains were selected to cover the complete 3ʹUTR diversity found in publicly available genome data. 3ʹUTR schemes are plotted to scale, highlighting the variable length and architectural organization of individual strains. Coloured boxes represent evolutionarily conserved, structured RNAs. Truncated sequences from incomplete sequencing are marked by an asterisk. The phylogenetic tree on the right has been computed from full genome nucleotide multiple sequence alignments of 28 TBEV strains. Identifiers show strain name, and lineage association where available, i.e. TBEV-FE Clusters I/II/III (C-I/C-II/C-III), the two Baikalean subtypes TBEV-Bkl-1 (Bkl-1), and TBEV-Bkl-2 (Bkl-2), and TBEV-Sib lineages Baltic (Bal), Zausaev (Zau), Vasilchenko (Vas), Bosnia (Bos), Obskaya (Obs). Accession numbers and 3ʹUTR lengths are listed in Supplementary Table S1.
