Abstract
Background:
Epstein-Barr virus (EBV) has been implicated in the development of breast cancer (BC) since 1995. It is classified into A/B genotypes, C/D subtypes, and F/f variants according to variations in its genome.
Aim:
To determine the distribution difference of EBV types between BC patients and healthy controls in Egypt and to detect the association between different EBV types and BC characteristics.
Methods:
Three hundred and sixty-two participants (142 BC patients and 220 controls) were enrolled in this study. All participants were screened for EBV infection by determination of viral-capsid-IgG antibodies in their sera. EBNA-1 gene was detected by PCR in tumor biopsies of seropositive patients and in peripheral blood mononuclear cells of controls. A/B genotyping of EBV was performed by nested-PCR targeting the EBNA-2 gene. C/D subtypes and F/f variants were identified by Restriction fragment length polymorphism at BamHI-I W1/I1 and BamHI-F regions of EBV genome, respectively.
Results:
Among 362 participants, 300(82.9%) were EBV-seropositive, including 120/142(84.5%) of the BC patients and 180/220(81.8 %) of the controls. EBNA-1 gene was positive in 54(45%) of seropositive BC patients and in 38(21.1%) of seropositive controls. There was a significant association of EBNA-1 gene with breast cancer (OR=3.05, 95%CI=1.84-5.07). Moreover, EBNA-1 gene positivity was significantly associated with the more aggressive tumors. Genotype-A and prototype-F were predominant among patients (90.4%, 100%, respectively) as well as among controls (91.7%, 100%, respectively) with no statistical significant association with BC risk. However, subtype-D was significantly more frequent in patients (95.6%) than in controls (64.7%) and was significantly associated with a higher BC risk as compared to subtype-C (OR=11.7, 95%CI=2.4-57.08). Subtype-D was significantly associated with higher grades tumors (100% among grade III), with progesteron receptor-negative tumors and with HER2-positive tumors (100% for each). The combined genotypes that significantly associated with BC risk were ADF (OR=4.9) and BDF (OR=5.5).
Conclusions:
Subtype-D of EBV could be the only EBV type implicated in BC development among Egyptian females and associated more with poor prognosis.
Key Words: Epstein, Barr virus, breast, cancer, RFLP, genotyping, Egypt
Introduction
Epstein-Barr virus (EBV), the etiological agent of infectious mononucleosis, is a Herpes virus that belongs to the Herpesviridae family. It is highly widespread, affecting more than 90% of the world’s population (Dowd et al., 2013).
EBV genome is 172,000 base pairs long and although it can code for more than 85 proteins, the well-known ones are few; Six EBV nuclear antigens (EBNAs 1, 2, 3A, 3B, and 3C, and EBNA-LP); three latent membrane proteins (LMPs 1, 2A, 2B), also known as latent genes , and small noncoding EBV-encoded RNAs (EBERs 1 and 2) (Kalla and Hammerschmidt, 2012).
The presence of polymorphisms in several regions of the EBV genome define different EBV types and variants, which have been shown to have a distinctive geographical distribution (Ayadi et al., 2006).
EBV is classified into two genotypes: genotype-A and genotype -B (sometimes known as type-1 and type-2). EBV Genotypes-A and-B were initially distinguished based on changes in the EBNA2 sequence ,which only shows 70% of gene homology and 54% of protein homology between genotype-A and genotype -B (Farrell, 2015). Although Palser and his colleagues observed that differences between genotype-A and genotype -B could be caused by variations in the sequences of EBNA2 and EBNA3A, EBNA3B, and EBNA3C genes, they found that the most frequent variance between genotype-A and genotype -B is the differences in the EBNA2 gene (Palser et al., 2015).
EBV BamHI-F and BamHI-W/ Bam HI-I boundary regions in the EBV genome have been identified as BamHI endonuclease restriction sites. Therefore, restriction fragment polymorphism (REFLP) analysis would help to further classify each of genotype-A and genotype-B into C/D subtypes and into F/f variants based on the presence or lack of BamHI restriction sites in the previously mentioned regions. Subtype-C is distinguished by the absence of a BamHI restriction site between the BamHI-W and BamHI-I fragments, while subtype-D has an extra BamHI site in the same region. In the BamHI-F fragments, prototype-F has no BamHI site, whereas variant-f has an extra BamHI site (Ayadi et al., 2007).
Diagnosis of EBV infection depends on a variety of tests, including nonspecific tests like the monospot test for heterophile antibodies, serological tests for detection of EBV- specific antibodies and molecular assays for detection of nucleic acid (Hess, 2004). Viral capsid antigen (VCA)-IgG, VCA-IgM, and EBNA-1 IgG are frequently required for the detection of EBV antibodies. VCA-IgG antibodies are superior to VCA-IgM in diagnosing past EBV infections. Furthermore, approximately 5–10% of healthy people infected with EBV never produce EBNA-IgG and this percentage is higher in immunodeficient people. Therefore, VCA-IgG antibodies are the most accurate single test for detection of past infection with EBV (Smatti et al., 2018). EBNA1 gene is constantly present in all EBV-infected cells. As a result, it is used for molecular detection of EBV infection (Sun et al., 2015).
Breast cancer (BC) is the most frequently diagnosed malignancy as well as the leading cause of cancer death among women, worldwide. In accordance to WHO estimates, BC would be diagnosed in 2.1 million women every year. According to reports in 2018, over 627,000 women died from the disease, accounting for almost 15% of all cancer-related fatalities among women, worldwide (Farahmand et al., 2019). BC is a complex disease with an aetiology that is poorly understood. Many traditional factors were identifid as risk factors for BC; family history, early menarche, late menopause, obesity and nulliparity (Kaminska et al., 2015).
In 1995, Labrecque and his colleagues were the first to identify the association between EBV and BC, and since then, many studies have confirmed the role of EBV in the development of BC (Labrecque et al., 1995). Recently, there has been an increasing interest to specify the association between different types of EBV and malignant diseases (Neves et al., 2017). However, the majority of these studies were focused on EBV-associated nasopharyngeal carcinoma (NPC) and EBV-associated gastric carcinoma, and they proposed that the pathological effects of EBV might vary according to the type of EBV strain (Corvalan et al., 2006; Chen et al., 2010).
To date, most of the researches that studied the relationship between EBV and breast carcinoma were interested in investigating whether or not there is an association between EBV infection and the development of BC, and none of these studies determined the association between different types of EBV and BC. Therefore, we investigated the variations in the distribution of different types, subtypes, and variants of EBV between BC women and healthy controls in Egypt and to clarify the relationship between specific EBV types and the histopathological features of BC.
Materials and Methods
Study subjects
This case-control study included 362 female participants; 142 breast cancer (BC) patients and 220 healthy controls. All patients and controls were recruited from the Oncology Center, Faculty of Medicine, Mansoura University (OCMU) in Egypt through the period from January 2019 to December 2020. The ages of all patients and controls were matched (p >0.05). Patients’ ages ranged from 37 to 79 years old with a median age of 54 years. The diagnosis of BC in all enrolled patients was based on clinical examination, radiological findings, and histopathological examination of tissue biopsy. Control subjects were selected from women who attended OCMU for annual mammography as part of a breast checkup and we excluded those with a history of BC and those with fibroadenomas.
Sample collection
Fresh tumor tissue and blood specimen were obtained from BC patients, whereas only blood specimen was obtained from the control group. The tissue sample from each tumor was divided into two parts; one part was fixed in formaldehyde and delivered to OCMU’s pathology laboratory for histopathological examination, while the other part was stored at -80°C to be used for PCR. The blood specimen was obtained via venipuncture and divided into two tubes; a plain tube for separation of serum and an EDTA tube for separation of peripheral blood mononuclear cells (PBMCs), both of which were kept at –80°C until use.
Study design
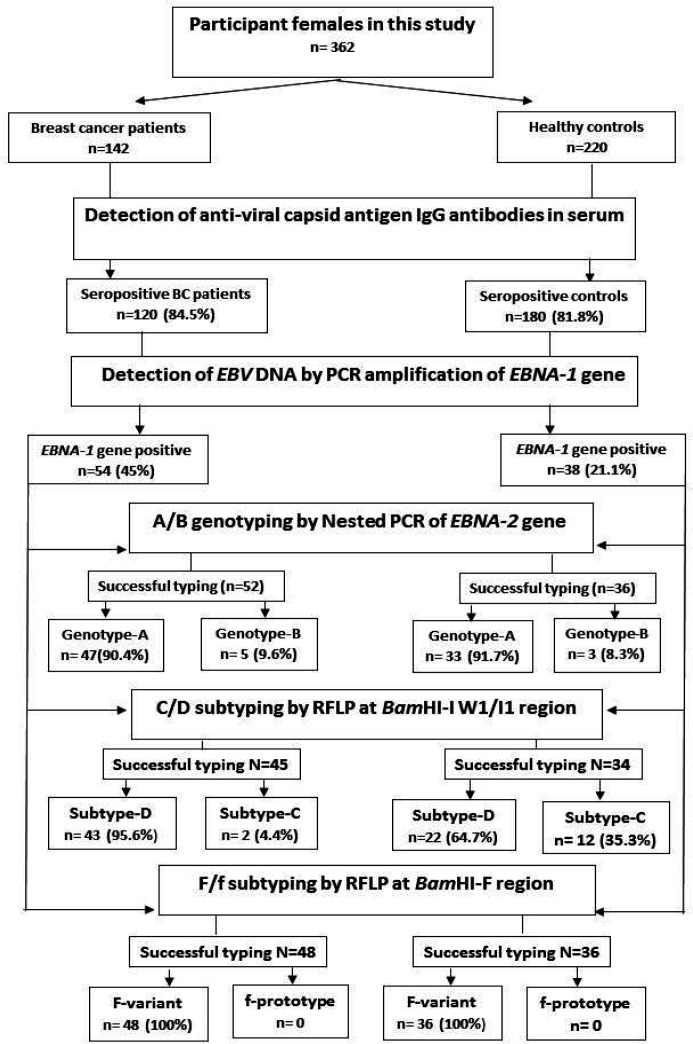
A Flowchart showing the workflow of this study is presented in Figure 1. All included BC patients and controls were screened for EBV infection by the detection of anti-viral capsid antigen (VCA) IgG antibodies in their sera. Then EBV seropositive subjects were investigated for the presence of EBV DNA by PCR amplification of the EBNA-1 gene; either in tumor biopsy of BC patients or in PBMCs of controls. Thereafter, A/B genotypes, C/D subtypes and F/f variants of EBV were identified in all EBV-DNA positive specimens by analysis of three regions in the EBV genome (EBNA-2 gene, BamHI-I W1/I1 and BamHI-F regions, respectively).
Figure 1.
Flowchart Indicating the Design and Results of the Present Study
Histopathological examination
Diagnosis of BC and determination of its histological type was achieved in OCMU’s pathology laboratory by examination of hematoxylin-eosin (H-E) stained slides prepared from tumor biopsies. Only tumor biopsies with cancerous cells of more than 60% were included in the study. Grading of the tumors was performed according to the Scarff Bloom Richardson classification (Bloom and Richardson, 1957). Detection of hormonal receptor status [estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor-2 (HER-2)] of all included tumor biopsies was done by immunohistochemical staining of slides cut from tissue microarray prepared blocks as described previously (Ahmed and Yussif, 2016).
Detection of EBV-viral capsid antigen (VCA) IgG antibodies
Screening of all included BC patients and controls for EBV infection was performed by detection of anti-VCA IgG antibodies by ELISA (Bio-Rad Medical Diagnostics, GmbH, Germany) following the manufacturer guidelines.
Polymerase chain reaction (PCR) amplification of EBNA-1 gene
Detection of EBV DNA was performed by PCR amplification of EBNA-1 gene. DNA was extracted from all specimens with serologically confirmed EBV infection (120 tumor biopsies from BC patients and 180 PMNCs from controls) using the QIAamp DNA Mini Kit (Qiagen, GmbH, Hilden, Germany), following the manufacturer’s tissue and blood protocols. Then EBV DNA was detected by PCR amplification of the EBNA-1 gene using specific primers listed in Table 1 according to the previously described PCR protocol (Hashimoto et al., 1995).
Table 1.
Target Genes and Their Primer Sequence
| Target gene | Primer sequence | Product size (bp) |
|---|---|---|
| EBNA-1 | F: 5'CACTTTAGAGCTCTGGAGGA-3' R: 5'TAAAGATAGCAGCAGCGCAG-3' |
112 |
| EBNA-2 | ||
| First round primers | ||
| EBNA-2A | F: 5'TGGAAACCCGTCACTCTC-3' | 801 |
| EBNA-2B | R: 5'TAATGGCATAGGTGGAATG-3' | |
| Second round primers | ||
| EBNA-2C | F: 5'AGGGATGCCTGGACACAAGA-3' | |
| EBNA-2D | R: 5'GCCTCGGTTGTGACAGAG-3' | Genotype-A: 250 |
| EBNA-2E | R: 5'TTGAAGAGTATGTCCTAAGG-3' | Genotype-B: 300 |
| BamHI-I W1/I1 region | F: 5'ACCTGCTACTCTTCGGAAAC-3' R: 5'TCTGTCACAACCTCACTGTC-3' |
Subtype-C: 205 Subtype-D: 130 + 75 |
| BamHI-F region | F: 5'TCCCAGCTGTTACCACATTC-3' R: 5'GGCAATGGGACGTCTTGTAA-3' |
Prototype-F:198 Variant-f: 127 + 71 |
bp, base pair; F, forward; R, reverse.
Detection of A/B genotypes of EBV
Nested PCR that targets EBNA-2 gene was used for genotyping of EBV into genotype-A and genotype-B (Ayee et al., 2020). In the first round of PCR amplification, a common region of EBNA-2 was amplified using EBNA-2A and EBNA-2B as forward and reverse primers, respectively. A PCR reaction was performed in a total volume of 12.5ul and the final concentrations of each component were as follows; 1X One TaqR Quick-Load 2X master mix with standard buffer (Bio Labs, UK), 0.5µM from each of the forward and reverse primers, and 0.5µg/µL of the extracted DNA. Amplification was achieved in a thermal cycler under the following conditions; 2min at 94°C for initial denaturation, then followed by 35 cycles of amplification (each cycle consisted of 60s at 94°C, 90s at 52°C and 4min at 72°C), followed by a cycle of 10min at 72°C for final extension.
In the 2nd round of nested PCR, the reaction volume was 12.5ul and the components were the same as in the 1st round except that 0.5ul of the amplified product from the 1st round was used as a template and three primers were used; EBNA-2C as a forward primer (common to genotype-A and genotype-B), EBNA-2D and EBNA-2E as reverse primers specific for genotype-A and genotype-B, respectively. The cycling conditions were as follows: An initial cycle of heat denaturation for 2min at 94°C, 35 amplification cycles (30s at 94°C, 60s at 52°C, and 2 min at 72°C), and a final extension cycle for 10min at 72°C. The resultant amplicon was visualized by electrophoresis on an agarose gel with 2% ethidium bromide. Samples were identified by the presence of their respective bands (250 bp for genotype-A and 300 bp for genotype-B).
Detection of C/D subtypes and F/f variants of EBV
Restriction fragment length polymorphism (RFLP) was used for classification of the EBV into C/D subtypes and F/f variants according to the existence of an extra restriction enzyme site at the BamHI-I W1/I1 and BamHI-F regions of the EBV genome, respectively. PCR was performed, followed by RFLP as previously described (Henry et al., 2001).
In brief, the reaction conditions for PCR amplification of each region were similar except for the use of forward and reverse primers specific for each region. The total reaction volume was 25ul consisting of 22.0µl of PCR master mix (1x), 1µl of each forward and reverse primer (0.5μM), and 1µl of DNA template. PCR cycles included an initial step of denaturation at 94°C for 5min followed by 40 amplification cycles; denaturation at 94ºC for 30s, annealing at 55°C for 30s and extension at 72°C for 1min , thereafter a step of final extension for 10min at 72°C. After PCR, the enzymatic digestion of the amplified product was performed in a total reaction volume of 20ul that contained 10µl of the amplified products, 2µl of reaction buffer, 1.5ul of distilled water, and 1.5µl of BamHI endonucleases (10U/ul) (Promega, Madison). This mixture was incubated at 37ºC for 2hs then followed by agarose gel (2%) electrophoresis of enzyme-digested BamHI-I W1/I1 region and BamHI-F region for determination of C/D subtypes and F/f variants, respectively. The product size of BamHI W1/I1 region after enzyme digestion is either 206 bp with subtype-C or 139bp and 67bp with subtype-D. For the products of enzyme digested BamHI F region, the presence of one band (199 bp) identified an F-variant whereas the presence of two bands (128bp and 71bp) indicated an f-variant.
Statistical analysis
The Statistical Package for Social Sciences (SPSS) version 22 was used to analyze the data. The qualitative data were expressed as frequencies and percentages. The differences in distribution of EBV types between tissue biopsies of BC patients and sera of the control group were compared by a univarite binary logistic regression test with the calculation of odds ratio (OR) and 95% confidence intervals (CIs). The difference between groups was considered statistically significant if the p-value was less than 0.05.
Results
Detection of anti-VCA IgG of EBV by ELISA
Screening of 142 BC patients and 220 healthy females for past exposure to EBV infection revealed that 120/142 (84.5%) of the cases and 180/220 (81.8%) of the controls were seropositive for anti-VCA IgG with no statistically significant difference between them (p=0.5), Table 2.
Table 2.
Frequency of EBV Infection in Breast Cancer Patients and Controls
| Control group No. (%) |
BC patients No. (%) |
P-value | OR (95% CI) | |
|---|---|---|---|---|
| Anti- VCA IgG (n) | 220 | 142 | ||
| Seronegative | 40 (18.2) | 22 (15.5) | Reference | |
| Seropositive | 180 (81.8) | 120 (84.5) | 0.507 | 1.2 (0.68-2.14) |
| EBNA-1 gene (n) | 180 | 120 | ||
| Negative | 142 (78.9) | 66 (55) | Reference | |
| Positive | 38 (21.1) | 54 (45) | <0.001 | 3.05 (1.84-5.07) |
EBV, Epstein-Barr virus; VCA, Viral capsid antigen; EBNA, Epstein Barr virus nuclear antigen; BC, breast cancer.
Detection of EBV DNA by PCR
EBV-DNA was detected in 54/120(45%) of BC tissue biopsies and in 38/180 (21.1%) of control PBMCs. There was a significant association between BC and EBV-DNA positivity (OR=3.05, 95%CI: 1.84-5.07, p<0.001), Table 2.
Genotyping of EBV
Analysis of A/B types of EBV
Successful amplification of the EBNA-2 gene was observed in 52/54 (96.3%) of BC patients and in 36/38 (94.7%) of controls. Nested PCR of the EBNA-2 gene revealed that genotype-A EBV was more frequently detected than genotype-B EBV in both BC patients (90.4% vs. 9.6%) and healthy control groups (91.7% vs. 8.3%). There was no statistically significant difference in the distribution of genotypes-A and -B between BC patients and the control group (p=0.8), Table 3.
Table 3.
EBV Typing in Breast Cancer Patients and Controls
| EBV types | Number (%) | P- value | OR (95%CI) | |
|---|---|---|---|---|
| Control group (n=38) |
BC patints (n=54) |
|||
| A/B genotypes | ||||
| Successful typing | 36 (94.7) | 52 (96.3) | ||
| A-genotype | 33 (91.7) | 47 (90.4) | Reference | |
| B-genotype | 3 (8.3) | 5 (9.6) | 0.837 | 1.102 (0.43-2.78) |
| C/D subtypes | ||||
| Successful typing | 34 (89.5) | 45 (83.3) | ||
| C-subtype | 12 (35.3) | 2 (4.4) | Reference | |
| D-subtype | 22 (64.7) | 43 (95.6) | 0.002 | 11.7 (2.4-57.1) |
| F/f variants | - | |||
| Successful typing | 36 (94.7) | 48 (88.9) | ||
| F-variant | 36 (100) | 48 (100) | ||
| f-prototype | 0 | 0 | ||
| Combined types of EBV | ||||
| Successful typinga | 32 (84.2) | 39 (72.2) | ||
| ACF | 9 (28.1) | 1 (2.6) | Reference | |
| ADF | 20 (62.5) | 33 (84.6) | 0.005 | 4.92 (1.61-14.99) |
| BDF | 2 (6.2) | 4 (10.3) | 0.024 | 5.54 (1.25-24.41) |
| BCF | 1 (3.1) | 1 (2.6) | 0.217 | 3.62 (0.47-27.55) |
EBV, Epstein-Barr virus; VCA, Viral capsid antigen; EBNA, Epstein Barr virus nuclear antigen; NA, not amplified gene; BC, breast cancer; a, Combined types of EBV were calculated among 39 cases and 32 controls who were successfully typed for all 3 investigated regions
Analysis of C/D subtypes of EBV
Amplification of the BamHI W1/I1 boundary region was successful in 45/54 (83.3%) of BC patients and in 34/38 (89.5%) of controls. Among 45 BC patients with successfully amplified BamHI W1/I1 region, subtype-D was detected more frequently than subtype-C (95.6% vs. 4.4%). Similarly, in the control group, subtype-D was more frequent than subtype-C (64.7% vs. 35.3%). There was a statistically significant difference in the distribution of subtypes-D and-C between BC patients and the control group (p=0.002). Subtype-D EBV was significantly more frequently associated with tissue biopsies from breast tumors (95.6%) than with PMNCs from controls (64.7%) (OR=11.7, 95%CI= 2.4-57.1), Table 3.
Analysis of F/f variants of EBV
The BamHI-F region was amplified successfully in 48/54 (88.9%) of BC tumor biopsies and in36/38 (94.7%) of controls. The successfully amplified EBV from all BC patients and controls was of F-variant, Table 3.
Combined Genotypes of EBV
A comparison of the combined genotypes of EBV between BC patients and controls revealed that the most predominant combination of EBV genotypes among BC patients as well as among the control group was ADF (84.6% vs. 62.5%, respectively). In BC patients, BDF was the second most frequent combined genotype (10.3%), followed by ACF and BCF (2.6% for each). Whereas in the control group, the second most frequent combined genotype was ACF (28.1%), followed by BDF (6.2%) and BCF (3.1%). The distribution of the EBV genotype combination between BC patients and controls was statistically significant (p=0.022). The combined genotypes that significantly associated with BC risk were ADF (OR= 4.9, 95%CI= 1.61-14.99) and BDF (OR= 5.5, 95%CI= 1.25-24.41), Table 3.
Association between EBV DNA positivity and tumor characteristics
Among 120 seropositive BC patients, there was a significant association between detection of EBV-DNA (EBNA-1 gene) and each of the large tumor size and higher tumor stage (P = 0.001 for each). Also, positivity of EBV-DNA was significantly associated with carcinomas of infiltrating ductal type (75%), and with tumors of high histological grade (71.4%, 50%, and 14.3% for grades III, II, and I, respectively). Moreover, the proportion of EBV DNA positive tumors was significantly higher in metastatic tumors (p=0.001). On the other hand, there was no observed association between EBV-DNA detection in BC tissues and steroid hormone receptors. However, EBV-DNA was detected significantly more frequently in HER2-positve tumors than in HER2-negative tumors (75% vs. 25%, p= 0.001), Table 4.
Table 4.
Association between EBV-DNA Positivity and Character of Tumors among 120 EBV Seropositive BC Patients
| Tumor characteristics |
No. (%) of cases | No. of EBV-DNA positive cases*/ total No of cases (%) | P value |
|---|---|---|---|
| Tumor size | 0.001 | ||
| Less than 2 cm | 33 (27.5) | 3/33 (9.1) | |
| More than 2 cm | 87 (72.5) | 51/87 (58.6) | |
| Tumor stage | 0.001 | ||
| Stage 1 | 21 (17.5) | 3/21 (14.3) | |
| Stage 2 | 51 (42.5) | 15/51 (29.4) | |
| Stage 3 | 48 (40) | 36/48 (75) | |
| Tumor grade | 0.001 | ||
| Grade 1 | 42 (35) | 6/42 (14.3) | |
| Grade 2 | 36 (30) | 18/36 (50) | |
| Grade 3 | 42 (35) | 30/42 (71.4) | |
| Tumor Histology | 0.001 | ||
| IDC | 102 (85) | 54/102 (52.9)) | |
| ILC | 18 (15) | 0 | |
| Metastasis | 0.001 | ||
| No | 111 (92.5) | 45/111 (40.5) | |
| Yes | 9 (7.5) | 9/9 (100) | |
| ER | 0.6 | ||
| Negative | 57 (47.5) | 27/57 (47.4) | |
| Positive | 63 (52.5) | 27/63 (42.9) | |
| Tumor characteristics |
No. (%) of cases | No. of EBV DNA positive cases*/ total No of cases (%) | |
| PR | |||
| Negative | 66 (55) | 30/66 (45.5) | |
| Positive | 54 (45) | 24/54 (44.4) | |
| HER2 | |||
| Negative | 72(60) | 18/72(25) | |
| Positive | 48(40) | 36/48(75) |
*Total number of EBV DNA-positive in this study was 54 of 120 seropositive positive BC patients; EBV, Epstein-Barr virus; IDC, Infiltrating ductal carcinoma; ILC, Invasive lobular carcinoma; PR, Progesterone receptor; ER, Estrogen receptor; HER2, Human epidermal growth factor receptor-2
Association between EBV genotypes and tumor characteristics
A/B genotypes of EBV did not show significant associations with any of tumor characteristics. Similarly, no association of statistical significance was observed between the C/D subtypes of EBV and either tumor size, tumor stage, tumor histology, presence of metastasis, or oestrogen receptor. However, the C/D subtypes of EBV from BC biopsies showed significant associations with tumor grade, progesterone receptor, and HER2- positive tumors. The proportion of subtype D positive BC biopsies increased significantly with tumour grade, from 86.7% for grade II to 100% for grade III. Although the D-subtype of EBV was detected more frequently in progesterone receptor-negative tumors (100%) than in progesterone receptor-positive tumors (86.7%), it was significantly more frequent in HER2-positive tumors than HER2- negative tumors (100% vs. 83.3%, p= 0.01), Table 5.
Table 5.
Association between Specific EBV Genotypes and Tumor Features
| Tumor characteristics |
A/B genotypes No. (%) (n=52) | D/C subtypes No. (%) (n=45) | ||||||
|---|---|---|---|---|---|---|---|---|
| No. (%) of cases |
Genotype- A | Genotype- B | P-value | Total No. (%) of cases | Subtype-D | Subtype-C | P-value | |
| Tumor size | 0.4 | 0.6 | ||||||
| less than 2 cm | 4 (7.7) | 4/4 (100) | 0 | 3 (6.7) | 3/3 (100) | 0 | ||
| more than 2 cm | 48 (92.3) | 43/48 (89.6) | 5/48 (10.4) | 42 (93.3) | 40/42 (95.2) | 2/42 (4.8) | ||
| Tumor stage | 0.2 | 0.4 | ||||||
| stage 1 | 3 (5.8) | 3/3 (100) | 0 | 3 (6.7) | 3/3 (100) | 0 | ||
| stage 2 | 16 (30.8) | 16/16 (100) | 0 | 15 (33.3) | 15/15 (100) | 0 | ||
| stage 3 | 33 (63.5) | 28/33 (84.8) | 5/33 (15.2) | 27 (60) | 25/27 (92.6) | 2/27 (7.4) | ||
| Tumor grade | 0.5 | 0.04 | ||||||
| Grade 1 | 5 (9.6) | 5/5 (100) | 0 | |||||
| Grade 2 | 17 (32.7) | 16/17 (94.1) | 1/17 (5.9) | 15 (33.3) | 13/15 (86.7) | 2/15 (13.3) | ||
| Grade 3 | 30 (57.7) | 26/30 (86.7) | 4/30 (13.3) | 30 (66.7) | 30/30 (100) | 0 | ||
| Tumor Histology | 0.7 | - | ||||||
| IDC | 51 (98.1) | 46/51 (90.2) | 5/51 (9.8) | 45 (100) | 43/45 (95.6) | 2/45 (4.4) | ||
| ILC | 1 (1.9) | 1/1 (100) | 0 | 0 | 0 | 0 | ||
| Metastasis | 0.8 | 0.4 | ||||||
| No | 43 (82.7) | 39/43 (90.7) | 4/43 (9.3) | 36 (80) | 34/36 (94.4) | 2/36 (5.6) | ||
| Yes | 9 (17.3) | 8/9 (88.9) | 1/9 (11.1) | 9 (20) | 9/9 (100) | 0 | ||
| ER | 0.6 | 0.07 | ||||||
| Negative | 26 (50) | 23/26 (88.5) | 3/26 (11.5) | 27 (60) | 27/27 (100) | 0 | ||
| Positive | 26 (10) | 24/26 (92.3) | 2/26 (7.7) | 18 (40) | 16/18 (88.9) | 2/18 (11.1) | ||
| PR | 0.8 | 0.04 | ||||||
| Negative | 29 (55.8) | 26/29 (89.7) | 3/29 (10.3) | 30 (66.7) | 30/30 (100) | 0 | ||
| Positive | 23 (44.2) | 21/23 (91.3) | 2/23 (8.7) | 15 (33.3) | 13/15 (86.7) | 2/15 (13.3) | ||
| HER2 | 0.7 | 0.01 | ||||||
| Negative | 18 (34.6) | 16/18 (88.9) | 2/18 (11.1) | 12 (26.7) | 10/12 (83.3) | 2/12 (16.7) | ||
| Positive | 34 (65.4) | 31/34 (91.2) | 3/34 (8.8) | 33 (73.3) | 33/33 (100) | 0 | ||
IDC, Infiltrating ductal carcinoma; ILC, Invasive lobular carcinoma; PR, Progesterone receptor; ER, Estrogen receptor; HER2, Human epidermal growth
Discussion
The presence of different polymorphisms in certain regions of the EBV genome resulted in the presence of different genotypes, subtypes, and variants of EBV (Ayadi et al., 2007). However, little is known about how EBV genetic diversity may affect EBV-associated disease (Palser et al., 2015). Moreover, up to date, there is a controversy whether these EBV variants are geographically related, or disease-associated. In 1995, the association of EBV with BC was reported but no study has been conducted in Egypt since then to investigate if there is a specific association of BC with certain types of EBV or if it is simply a geographical distribution.
In this study, the seroprevalence of EBV was 84.5% in BC patients and was 81.8% in the control group with no statistical significant difference. This is consistent with the high prevalence of EBV in the world. It is estimated that more than 90% of the world’s population is EBV-seropositive (Tzellos and Farrell, 2012). Smatti et al., (2017) reported that the EBV seroprevalence rate among healthy blood donors was 100% in Egyptians and Pakistanis, 98.3% in Indians, 96.8% in Syrians, and 97.8% in Qataris.
Although this study detected a high seroprevalence of EBV among all study participants, the latent infection with EBV (as detected by molecular detection of the EBNA-1 gene) was significantly higher in BC tumor biopsies (45%) as compared to PMNCs of the control group (21.1%). This is in line with two previous Egyptian studies that detected EBV infection in 35.3% and 25% of breast carcinomas (Mohamed et al., 2007; Fawzy et al., 2008). Additionally, using PCR as a method of EBV detection, the prevalence of EBV infection among BC patients in various regions of the world was about 10–50% (Perkins et al., 2006; Lorenzetti et al., 2010). Other studies, on the other hand, failed to detect EBV among BC patients (Lespagnard et al., 1995; Kijima et al., 2001). This discrepancy in the results could be attributed to a variety of factors, such as geographical variation, differences in EBV infection detection methods, specimen type, use of different genes in the EBV genome for PCR, and the inclusion of different histological types of BC. However, studies that used biopsy specimens and used nested PCR as a method for detection showed less heterogeneity (Farahmand et al., 2019).
In this case-control study, EBV-DNA positivity increased the risk of BC development by about threefold (OR = 3.05). This is similar to two recent meta-analysis that reported that the risk of BC association with EBV infection is 3.84 and 4.74 folds higher as compared to controls (Bae and Kim, 2016; Farahmand et al., 2019).
In the current study, in an attempt to clarify if there is an association between BC and specific types of EBV in Egypt, we detected the distribution frequency of different EBV types obtained from tumor tissues of BC women and compared them with those obtained from PMNCs of healthy non-BC females.
Regarding the distribution of A/B genotypes of EBV among the studied BC patients and healthy controls, we found that most of the EBV detected in both BC tumor biopsies and PBMNCs of the controls were of genotype-A (90.9% and 91.7%, respectively, p= 0.8). The high prevalence of genotype-A among the participants of this study is consistent with its geographical distribution. Genotype-A is the most prevalent genotype of EBV in the world, predominantly in the United States, France, Germany, North Africa, Turky, and Asia. Whereas, genotype of EBV predominates more in Central Africa and New Guinea (Ibrahim et al., 2010). In this study, the distribution difference of genotype-A and gentype-B was statistically insignificant between BC patients and the control group. As a result, it appears that neither genotype-A nor genotype-B has a preferential association with BC in Egypt. In the same context, the association of A/B genotypes of EBV with other malignant diseases such as NPC seems to be variable worldwide. Tamura et al (1993) reported no association of A/B genotypes of EBV with NPC in Japan, as they were predominant in patients as well as in general populations. Klemenc et al., (2006) and Ayadi et al., (2007), reported a high incidence of genotype-A in Slovenian and Tunisian patients with NPC, respectively. Ayee et al., (2020) identified genotype-B as the virulent genotype in Ghana and associated it with the likelihood of NPC development in Ghanaian patients.
Regarding the polymorphism in the BamHI-I W1/I1 region. This study demonstrated that the frequency of subtype-D EBV is significantly higher in BC tumor biopsies (95.6%) as compared to controls (64.7%) (OR =11.7, 95%CI = 2.4-57.1, P =0.002). Similarly, Zhang et al., (2017) found a significant association between subtype-D and the risk of BC among Chinese populations (OR = 2.86), suggesting that subtype-D may play a role in the pathogenesis of BC. It is worth noting that the association of EBV subtype-D with other malignant diseases has previously been demonstrated; Abdel Hamid et al., (1992) discovered that subtype-D is detected more frequently in Egyptian NPC than subtype-C. Similarly, it was reported that EBV of subtype-D was associated with NPC in each of Slovenian patients (62.5%), Tunisian population (98%) and Northern China (32.2%) (Cui et al., 2011). Moreover, subtype-D was found to be associated not only with NPC but also with gastric carcinoma in Iranian population (90%), in Southern Tunisian population (100%), in Latin American population (85%) (Corvalan et al., 2006; Abdirad et al., 2007; BenAyed-Guerfali et al., 2011).
The genetic evidence that C/D subtypes of EBV could be related to the development of tumors is that some EBV genes, such as BamH1-A Rightward Frame-1 (BARF1) and Latent Membrane Protein-2A are present near the C/D locus. These genes are involved in transformation and immortalization during the tumorigenic process (Corvalan et al., 2006). However, the precise mechanisms need more exploration.
Regarding the polymorphism at the BamHI-F region, we detected that EBV from all BC tumor biopsies and control PMNCs was of prototype-F. Therefore, the association of prototype-F with Egyptian BC is not likely. It was proposed that f-variant is only a geographic polymorphism because the EBV that was detected in Egyptian, Algerian, and Tunisian NPCs was of prototype-F, whereas the EBV that was detected in Asian NPCs was of variant-f (Cui et al., 2011). However, Zhou et al., (2001) reported that variant-f is found only in Chinese NPCs and not in Chinese HD patients. Therefore, the hypothesis that the f-variant is a geographically restricted polymorphism needs further investigation.
In the present study, ADF was the most prevalent combination of EBV genotypes among BC patients as well as among control subjects. Likewise, Klemenc et al., (2006) found that ADF is the most predominant combination of EBV genotypes in Slovenian patients with NPC as well as in the healthy population. Additionally, the risk of BC was significantly higher with each of ADF (OR=4.92) and BDF (OR=5.54) combinations and not significantly increased with BCF (P=0.217). Therefore, our findings could point to the implication of specific combinations of EBV genotypes in the development of BC in Egypt. Consequently, our results are not consistent with the hypothesis suggested by Khanim et al (1996) that EBV strains are geographically determined rather than disease-associated.
As regards the association between EBV and the characteristics of BC. EBV was detected more frequently in BC of larger size, higher stage, and higher grade. These findings are similar to that made previously by Murray et al., (2003) and Mohamed et al., (2007) who found that EBV is significantly associated with the more aggressive tumors. Other studies, however, debate our findings as they did not find any statistically significant association between EBV positivity and either grade, stage, or metastasis of the tumor (Richardson et al., 2015; Ahmed and Yussif, 2016; El-Naby et al., 2017).
In this study, there was no demonstrable significant association between EBV positivity and the steroid-hormone receptor. This agrees with many previous studies (Tiwawech et al., 2008; Ahmed and Yussif, 2016; Bae and Kim, 2016; Preciado et al., 2005). On the contrary, other investigators have shown that EBV is significantly associated with steroid hormone receptor-negative tumors (Bonnet et al., 1999; Murray et al., 2003; El-Naby et al., 2017).
We found that 75% of EBV-DNA positive BC biopsies are also reactive to HER2 expression. This is close to the result reported by Mohamed et al., (2007) who reported that 55.9% of DNA positive cases were HER2 positive. This finding correlates with the recognition that EBV could transform epithelial cells of the breast into malignancy via stimulating the HER2/HER3 signalling pathways (Szostek et al., 2009). Furthermore, our findings point to some extent that subtype-D of EBV could be the only EBV type that might be implicated in the development of BC with poor prognosis because it was the only type that showed more association with tumor of higher grade and with progesterone receptor-free tumors (p= 0.04).
In Conclusion, to the best of our knowledge, this is the first study in Egypt that provides baseline data on the prevalence of different EBV types in BC patients as well as in healthy controls. Overall, this study found that the D-subtype of EBV is BC-associated. Whereas, A/B genotypes and F/f variants of EBV are not associated with BC. Additionally, specific combinations of EBV genotypes (ADF and BDF) were BC- associated. Moreover, our findings indicated that EBV could have an impact on the histological criteria of BC and thus could have a role in prognosis and in optimizing the therapeutic strategies for BC. Nevertheless, our results are considered a platform for any future studies with a larger number of patients with BC in Egypt as well as in other parts of the world to investigate whether variations in the distribution of different types of EBV are geographically related or BC related.
Author Contribution Statement
Mashaly M: Contributes to the concept and design of the study, performed the laboratory work concerned with EBV diagnosis (ELISA, PCR, REFLP), wrote the original draft of the manuscript, revised statistical analysis and interpreted the obtained results. Ghorab D: performed the histopathological part of the study for breast cancer diagnosis. Hegazy M, Abdelkhalek M and Gaballah K: Contributed to the clinical examination of all participants and collection of the study samples and data. Elzehery R: Contributes to the concept and design of the study, performed the laboratory work concerned with EBV diagnosis (ELISA, PCR, REFLP). All authors read and revised the manuscript.
Acknowledgements
All authors would like to thank all the patients &staff for their participation and cooperation in this work.
Ethical approval
The present study received an ethical approval from Ethical Committee of Faculty of Medicine –Mansoura University (MFM-Institutional Research Board).
Availability of data
All data of this study are available on request in SPSS file.
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Abdel-Hamid M, Chen JJ, Constantine N, Massoud M, Raab-Traub N. EBV strain variation: geographical distribution and relation to disease state. Virology. 1992;190:168–5. doi: 10.1016/0042-6822(92)91202-6. [DOI] [PubMed] [Google Scholar]
- Abdirad A, Ghaderi-Sohi S, Shuyama K, et al. Epstein–Barr virus associated gastric carcinoma: A report from Iran in the last four decades. Diagn Pathol. 2007;2:25. doi: 10.1186/1746-1596-2-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed RA, Yussif SM. Immunohistochemical detection of human cytomegalovirus, Epstein-Barr virus and human papillomavirus in invasive breast carcinoma in Egyptian women: A tissue microarray study. J Solid Tumors. 2016;6:8–16. [Google Scholar]
- Ayadi W, Feki L, Khabir A, et al. Polymorphism analysis of Epstein-Barr virus isolates of nasopharyngeal carcinoma biopsies from Tunisian patients. Virus Genes. 2007;34:137–5. doi: 10.1007/s11262-006-0051-2. [DOI] [PubMed] [Google Scholar]
- Ayee R, Ofori MEO, Tagoe EA, et al. Genotypic Characterization of Epstein Barr Virus in Blood of Patients with Suspected Nasopharyngeal Carcinoma in Ghana. Viruses. 2020;12:766. doi: 10.3390/v12070766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae J, Kim E H. Epstein-Barr Virus Infection and Risk of Breast Cancer: An Adaptive Meta-Analysis for Case-Control Studies. Arch Clin Infect Dis. 2016;11:e34806. [Google Scholar]
- BenAyed-Guerfali D, Ayadi W, Miladi-Abdennadher I, et al. Characteristics of Epstein Barr virus variants associated with gastric carcinoma in Southern Tunisia. Virol J. 2011;8:500. doi: 10.1186/1743-422X-8-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom HJ, Richardson WW. Histological grading and prognosis in breast cancer; a study of 1409 cases of which 359 have been followed for 15 years. Br J Cancer. 1957;11:359–7. doi: 10.1038/bjc.1957.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet M, Guinebretiere JM, Kremmer E, et al. Detection of Epstein-Barr virus in invasive breast cancers. J Natl Cancer Inst. 1999;91:1376–81. doi: 10.1093/jnci/91.16.1376. [DOI] [PubMed] [Google Scholar]
- Chen JN, Ding YG, Feng ZY, et al. Association of distinctive Epstein-Barr virus variants with gastric carcinoma in Guangzhou, southern China. J Med Virol. 2010;82:658–7. doi: 10.1002/jmv.21731. [DOI] [PubMed] [Google Scholar]
- Corvalan A, Ding S, Koriyama C et al. Association of a distinctive strain of Epstein-Barr virus with gastric cancer. Int J Cancer. 2006;118:1736–2. doi: 10.1002/ijc.21530. [DOI] [PubMed] [Google Scholar]
- Cui Y, Wang Y, Liu X, et al. Genotypic analysis of Epstein-Barr virus isolates associated with nasopharyngeal carcinoma in Northern China. Intervirology. 2011;54:131–8. doi: 10.1159/000319632. [DOI] [PubMed] [Google Scholar]
- Dowd JB, Palermo T, Brite J, McDade TW, Aiello A. Seroprevalence of Epstein-Barr virus infection in U S children ages 6-19, 2003-2010. PLoS One. 2013;8:e64921. doi: 10.1371/journal.pone.0064921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Naby NEH, Mohamed HH, Goda AM, Mohamed AE. Epstein-Barr virus infection and breast invasive ductal carcinoma in Egyptian women: A single center experience. J Egypt Natl Canc Inst. 2017;29:77–2. doi: 10.1016/j.jnci.2017.02.002. [DOI] [PubMed] [Google Scholar]
- Farahmand M, Monavari SH, Shoja Z, et al. Epstein-Barr virus and risk of breast cancer: a systematic review and meta-analysis. Future Oncol. 2019;15:2873–5. doi: 10.2217/fon-2019-0232. [DOI] [PubMed] [Google Scholar]
- Farrell PJ. Epstein-Barr virus strain variation. Curr Top Microbiol Immunol. 2015;390:45–9. doi: 10.1007/978-3-319-22822-8_4. [DOI] [PubMed] [Google Scholar]
- Fawzy S, Sallam M, Awad NM. Detection of Epstein-Barr virus in breast carcinoma in Egyptian women. Clin Biochem. 2008;41:486–2. doi: 10.1016/j.clinbiochem.2007.12.017. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y, Nawata Y, Kurasawa K, et al. Investigation of EB virus and cytomegalovirus in rapidly progressive interstitial pneumonitis in polymyositis/dermatomyositis by in situ hybridization and polymerase chain reaction. Clin Immunol Immunopathol. 1995;77:298–6. doi: 10.1006/clin.1995.1156. [DOI] [PubMed] [Google Scholar]
- Henry S, Sacaze C, Berrajah L, et al. In nasopharyngeal carcinoma-bearing patients, tumors and lymphocytes are infected by different Epstein-Barr virus strains. Int J Cancer. 2001;91:698–4. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1110>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Hess RD. Routine Epstein-Barr virus diagnostics from the laboratory perspective: still challenging after 35 years. J Clin Microbiol. 2004;42:3381–7. doi: 10.1128/JCM.42.8.3381-3387.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim HA, Menasce LP, Pomplun S. Epstein-Barr virus (EBV) genotypes among human immunodeficiency virus (HIV)-related B-cell lymphomas and B-cell post-transplant lymphoproliferative disorders (B-PTLD)--late-onset lymphomas, especially in the HIV setting, are associated with type-B-EBV. Eur J Haematol. 2010;85:227–30. doi: 10.1111/j.1600-0609.2010.01460.x. [DOI] [PubMed] [Google Scholar]
- Kalla M, Hammerschmidt W. Human B-cells on their route to latent infection-early but transient expression of lytic genes of Epstein-Barr virus. Eur J Cell Biol. 2012;91:65–9. doi: 10.1016/j.ejcb.2011.01.014. [DOI] [PubMed] [Google Scholar]
- Kaminska M, Ciszewski T, Lopacka-Szatan K, Miot- la P, Staros- lawska E. Breast cancer risk factors. Prz Menopauzalny. 2015;14:196–2. doi: 10.5114/pm.2015.54346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanim F, Yao QY, Niedobitek G, et al. Analysis of Epstein-Barr virus gene polymorphisms in normal donors and in virus-associated tumors from different geographic locations. Blood. 1996;88:3491–1. [PubMed] [Google Scholar]
- Kijima Y, Hokita S, Takao S, et al. Epstein-Barr virus involvement is mainly restricted to lymphoepithelial type of gastric carcinoma among various epithelial neoplasms. J Med Virol. 2001;64:513–8. doi: 10.1002/jmv.1079. [DOI] [PubMed] [Google Scholar]
- Klemenc P, Marin J, Soba E, et al. Distribution of Epstein-Barr virus genotypes in throat washings, sera, peripheral blood lymphocytes and in EBV positive tumor biopsies from Slovenian patients with nasopharyngeal carcinoma. J Med Virol. 2006;78:1083–90. doi: 10.1002/jmv.20666. [DOI] [PubMed] [Google Scholar]
- Labrecque LG, Barnes DM, Fentiman IS, Griffin BE. Epstein-Barr virus in epithelial cell tumors: a breast cancer study. Cancer Res. 1995;55:39–5. [PubMed] [Google Scholar]
- Lespagnard L, Cochaux P, Larsimont D, et al. Absence of Epstein-Barr virus in medullary carcinoma of the breast as demonstrated by immunophenotyping, in situ hybridization and polymerase chain reaction. Am J Clin Pathol. 1995;103:449–52. doi: 10.1093/ajcp/103.4.449. [DOI] [PubMed] [Google Scholar]
- Lorenzetti MA, De Matteo E, Gass H, et al. Characterization of Epstein Barr virus latency pattern in Argentine breast carcinoma. PLoS One. 2010;5:e13603. doi: 10.1371/journal.pone.0013603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed WS, Mohamed MA, Omar MM. Possible involvement of Epstein-barr Virus (EBV) in pathogenesis and prognosis of female breast infiltrating duct carcinoma: clinicopathological, immunohistochemical and molecular study. Egypt J Med Microbiol. 2007;16:403–4. [Google Scholar]
- Murray PG, Lissauer D, Junying J. Reactivity with a monoclonal antibody to EpsteinBarr virus (EBV) nuclear antigen 1 defines a subset of aggressive breast cancers in the absence of the EBV genome. Cancer Res. 2003;63:2338–3. [PubMed] [Google Scholar]
- Neves M, Marinho-Dias J, Ribeiro J, Sousa H. Epstein-Barr virus strains and variations: Geographic or disease-specific variants? J Med Virol. 2017;89:373–7. doi: 10.1002/jmv.24633. [DOI] [PubMed] [Google Scholar]
- Palser AL, Grayson NE, White RE, et al. Genome diversity of Epstein-Barr virus from multiple tumor types and normal infection. J Virol. 2015;89:5222–7. doi: 10.1128/JVI.03614-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins RS, Sahm K, Marando C, et al. Analysis of Epstein-Barr virus reservoirs in paired blood and breast cancer primary biopsy specimens by real time PCR. Breast Cancer Res. 2006;8:R70. doi: 10.1186/bcr1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preciado MV, Chabay PA, De Matteo EN, et al. Epstein-Barr virus in breast carcinoma in Argentina. Arch Pathol Lab Med. 2005;129:377–1. doi: 10.5858/2005-129-377-EVIBCI. [DOI] [PubMed] [Google Scholar]
- Richardson AK, Currie MJ, Robinson BA, et al. Cytomegalovirus and Epstein-Barr virus in breast cancer. PLoS One. 2015;10:e0118989. doi: 10.1371/journal.pone.0118989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smatti MK, Al-Sadeq DW, Ali NH, et al. Epstein-Barr virus epidemiology, serology, and genetic variability of LMP-1 oncogene among healthy population: An Update. Front Oncol. 2018;13:8–211. doi: 10.3389/fonc.2018.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Zhao Z, Liu S, et al. Sequence Variation Analysis of Epstein-Barr Virus Nuclear Antigen 1 Gene in the Virus Associated Lymphomas of Northern China. PLoS One. 2015;10:e0140529. doi: 10.1371/journal.pone.0140529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostek S, Zawilinska B, Kopec J, Kosz-Vnenchak M. Herpesviruses as possible cofactors in HPV-16-related oncogenesis. Acta Biochim Pol. 2009;56:337–2. [PubMed] [Google Scholar]
- Tamura S, Kunimoto M, Tabata T, Yoshie O. Genotypic analysis of Epstein-Barr virus associated with nasopharyngeal carcinoma of Japanese patients. Jpn J Cancer Res. 1993;84:246–9. doi: 10.1111/j.1349-7006.1993.tb02863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwawech D, Srivatanakul P, Karalak A, Ishida T. Association between EBNA2 and LMP1 subtypes of Epstein-Barr virus and nasopharyngeal carcinoma in Thais. J Clin Virol. 2008;42:1–6. doi: 10.1016/j.jcv.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Tzellos S, Farrell PJ. Epstein-barr virus sequence variation-biology and disease. Pathogens. 2012;1:156–4. doi: 10.3390/pathogens1020156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Wang MY, Wei XL, et al. Associations of Epstein-Barr Virus DNA in PBMCs and the Subtypes with Breast Cancer Risk. J Cancer. 2017;8:2944–9. doi: 10.7150/jca.20330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Sandvej K, Li P, et al. Epstein-Barr virus gene polymorphisms in Chinese Hodgkin’s disease cases and healthy donors: Identification of three distinct virus variants. J Gen Virol. 2001;1:1157–7. doi: 10.1099/0022-1317-82-5-1157. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data of this study are available on request in SPSS file.