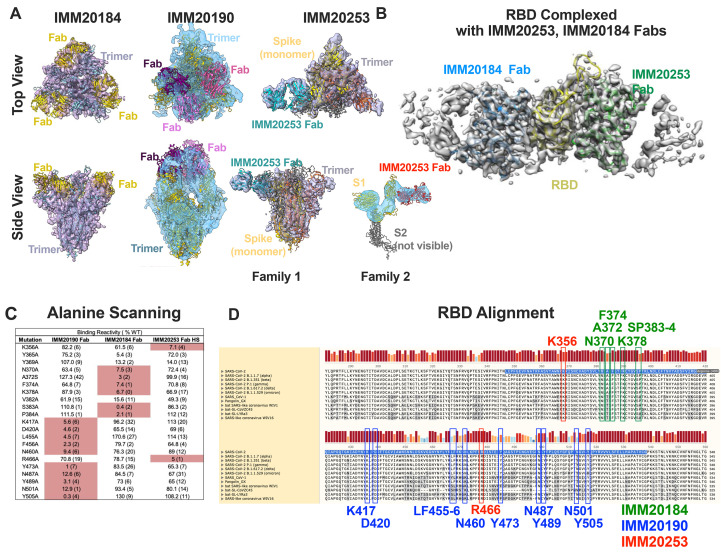
Fig. 1. IMM20184, IMM190 and IMM20253 antibodies bind to conserved epitopes and disrupt Spike Trimer.
(A) 3D reconstruction of Cryo-EM images of the Spike Trimer complex with either IMM20184 (left), IMM20190 (middle) or IMM20253 (right) Fabs. Top-to bottom view (top) and side view (bottom) are shown. IMM20253 Fab binding to Trimer results in two families, Family 1 and 2. Models PDB:7E8C, PDB:6XLU, PDB:6XM5 or PDB:7NOH were fit to density in Chimera for the Spike Trimer and PDB:6TCQ for the Fabs (see Supp Fig. 1A-E for details). (B) 3D reconstruction of Cryo-EM images of RBD complexed with simultaneously bound Fabs of IMM20184 and IMM20253. Fab model PDB:1M71 was fit to density in Chimera (see Sup Fig. 1F-J for details. (C) Critical residues of antibody epitopes identified as binding pattern to a library of single-point RBD mutants expressed on the cell surface. (D) Alignment of Spike protein sequences from current and prior CDC VOCs, SARS-CoV-1 and closely related coronaviruses. Critical residues of IMM20190, IMM20184 and IMM20253 epitopes are shown in blue, green and red. Highlighted sequence indicates RBD.