Abstract
Background
COVID-19 is a multi-system disorder with high variability in clinical outcomes among patients who are admitted to hospital. Although some cytokines such as interleukin (IL)-6 are believed to be associated with severity, there are no early biomarkers that can reliably predict patients who are more likely to have adverse outcomes. Thus, it is crucial to discover predictive markers of serious complications.
Methods
In this retrospective cohort study, we analysed samples from 455 participants with COVID-19 who had had a positive SARS-CoV-2 RT-PCR result between April 14, 2020, and Dec 1, 2020 and who had visited one of three Mayo Clinic sites in the USA (Minnesota, Arizona, or Florida) in the same period. These participants were assigned to three subgroups depending on disease severity as defined by the WHO ordinal scale of clinical improvement (outpatient, severe, or critical). Our control cohort comprised of 182 anonymised age-matched and sex-matched plasma samples that were available from the Mayo Clinic Biorepository and banked before the COVID-19 pandemic. We did a deep profiling of circulatory cytokines and other proteins, lipids, and metabolites from both cohorts. Most patient samples were collected before, or around the time of, hospital admission, representing ideal samples for predictive biomarker discovery. We used proximity extension assays to quantify cytokines and circulatory proteins and tandem mass spectrometry to measure lipids and metabolites. Biomarker discovery was done by applying an AutoGluon-tabular classifier to a multiomics dataset, producing a stacked ensemble of cutting-edge machine learning algorithms. Global proteomics and glycoproteomics on a subset of patient samples with matched pre-COVID-19 plasma samples was also done.
Findings
We quantified 1463 cytokines and circulatory proteins, along with 902 lipids and 1018 metabolites. By developing a machine-learning-based prediction model, a set of 102 biomarkers, which predicted severe and clinical COVID-19 outcomes better than the traditional set of cytokines, were discovered. These predictive biomarkers included several novel cytokines and other proteins, lipids, and metabolites. For example, altered amounts of C-type lectin domain family 6 member A (CLEC6A), ether phosphatidylethanolamine (P-18:1/18:1), and 2-hydroxydecanoate, as reported here, have not previously been associated with severity in COVID-19. Patient samples with matched pre-COVID-19 plasma samples showed similar trends in muti-omics signatures along with differences in glycoproteomics profile.
Interpretation
A multiomic molecular signature in the plasma of patients with COVID-19 before being admitted to hospital can be exploited to predict a more severe course of disease. Machine learning approaches can be applied to highly complex and multidimensional profiling data to reveal novel signatures of clinical use. The absence of validation in an independent cohort remains a major limitation of the study.
Funding
Eric and Wendy Schmidt.
Introduction
The ongoing COVID-19 pandemic is a public health emergency that would benefit from a systematic investigation of molecular alterations in patients to predict outcomes for improving clinical care. Currently, no conclusive cytokine-based treatments exist, although there are indications that targeted therapy could improve outcomes. Although interleukin (IL)-6 elevation has been associated with severe disease, its predictive capacity for clinical outcomes is poor. IL-6 receptor blockers such as tocilizumab have shown conflicting results in clinical trials,1, 2, 3 and have suggested substantial benefits of IL-6 blockade in select patients; however, we have no ability to identify these patients clinically. Similarly, IL-1 blockade with high-dose anakinra in patients with COVID-19 has shown inconsistent results.4 Tumour necrosis factor (TNF) inhibitors have been associated with less severe clinical phenotypes but clinical trial data are sparse.5 Lenzilumab has shown promise as a granulocyte-macrophage colony-stimulating factor (GM-CSF) inhibitor, whereas otilimab failed to show similar benefit.6 Ultimately, all of these results point to diverse inflammatory phenotypes that need to be better understood to individualise therapy.
Research in context.
Evidence before this study
Cytokine storm has been shown to be associated with severe COVID-19, and proinflammatory cytokines have been suggested as markers. We searched PubMed and Google Scholar for articles published from Jan 1, 2020, to Jan 31, 2022, without any language restrictions. We used keywords including “COVID-19”, “SARS-CoV-2”, “multi-omics”, “cytokines”, “lipidomics”, “metabolomics”, “proteomics”, “predictive biomarker”, and “machine learning”. Many articles reported proinflammatory cytokines of the innate immune response, such as interleukin(IL)-1, IL-6, and tumour necrosis factor, among cytokines and chemokines measured in plasma or serum as early markers of unfavourable outcomes. At least six recent studies focused on measuring the concentrations of hundreds of circulatory proteins, and six studies profiled lipids or metabolites in smaller cohorts (n<100). Only three studies included an integrated multiomics analysis of proteins, lipids, and metabolites across samples, although they had an inadequate number of controls, sampled patients at different stages of hospitalisation, and did not include severity criteria.
Added value of this study
To our knowledge, this study of 637 individuals, including 455 samples from patients positive for COVID-19 at different levels of severity as defined by the WHO ordinal scores of clinical improvements, represents the largest profiling of plasma samples from such a population. Coupling multiomics analysis with a machine-learning-based stratification revealed an early signature of COVID-19 severity that was validated in a held-out dataset. Our study led to the discovery of multiomic signatures in the early course of disease that are better predictors of adverse outcomes than the classic cytokine storm panel of cytokines.
Implications of all the available evidence
Availability of deep, unbiased profiling data using multiomic platforms is crucial for the discovery of predictors of disease severity. Along with existing markers, these novel markers could lead to better management of patients and reduce mortality of COVID-19.
We sought to combine concentrations of specific cytokines with other omics data for an individualised approach to predict outcomes in patients with COVID-19. Several studies have investigated cytokine, proteomic, metabolomic, and lipidomic changes.7, 8, 9, 10, 11 However, only a few have combined a multiomics approach with machine learning for an integrated analysis.10, 11 Notably, although some potential markers for COVID-19 have been suggested, they suffer from several limitations: small sample sizes (<100 participants in most studies); plasma samples that have been collected after hospital admission or after occurrence of serious clinical outcomes; group comparisons without adequate controls or without mild COVID-19 cases; disparate definitions of severity; and participants without matched samples before COVID-19 infection. In this study, we aim to develop a machine learning-based prediction model to discover a signature predictive of severe disease in patients with COVID-19.
Methods
Study design and participants
This was a retrospective cohort study of 455 participants with COVID-19 who had tested positive for SARS-CoV-2 by RT-PCR between April 14, 2020, and Dec 1, 2020 and who had visited one of three Mayo Clinic sites in the USA (Minnesota, Arizona, or Florida) in the same time period. Blood samples were obtained from each of these patients. Additionally, we used 182 anonymised age-matched and sex-matched control plasma samples that were available from the Mayo Clinic Biorepository and banked before the COVID-19 pandemic.12 As the control cohort is comprised of patients who have medical appointments at Mayo Clinic, they have the typical comorbidities of patients in the population that presents to Mayo Clinic. Four patient subgroups were defined using the WHO ordinal scale of clinical improvement (OSCI): 182 control cases (WHO OSCI=0), 183 outpatients with COVID-19 (WHO OSCI=1–2), 139 patients with severe COVID-19 (WHO OSCI=3–4), and 133 patients with critical COVID-19 (WHO OSCI=5–8). All plasma samples used in the study were obtained with informed consent from each subject. This study was approved by the institutional review board (IRB protocol 20–005760) of Mayo Clinic in accordance with the Declaration of Helsinki.
Procedures
WHO OSCI was assigned by attending physicians before the study on the basis of the patient's highest WHO OSCI score during COVID-19 illness (table 1 ; appendix 1 p 7). 190 (70%) of 272 samples were obtained from case participants within 3 days of hospitalisation or before hospitalisation. Details on plasma collection are provided in appendix 1 (p 2). The time of baseline plasma sample collection of participants with severe and critical COVID-19 is shown in appendix 1 (p 7). 24 (5%) of 455 patients had matched pre-COVID-19 plasma samples available in the Mayo Clinic Biorepository (12 outpatients, six patients with severe COVID-19, and six patients with critical COVID-19; appendix 1 p 7).
Table 1.
Baseline characteristics of the study cohort
| Control group (n=182) | Outpatients (n=183) | Patients with severe COVID-19 (n=139) | Patients with critical COVID-19 (n=133) | p value | |||
|---|---|---|---|---|---|---|---|
| Age, years | .. | .. | .. | .. | <0·0001 | ||
| Median | 58·8 (46·0–71·8) | 47·9 (31·2–60·6) | 62·1 (49·0–73·9) | 63·1 (52·3–74·0) | .. | ||
| Range | 21·0–98·0 | 18·5–80·9 | 18·2–101·9 | 24·6–96·4 | .. | ||
| Sex | .. | .. | .. | .. | 0·0019* | ||
| Female | 98 (54%) | 111 (61%) | 69 (50%) | 52 (39%) | .. | ||
| Male | 84 (46%) | 72 (39%) | 70 (50%) | 81 (61%) | .. | ||
| Body-mass index | .. | .. | .. | .. | 0·31† | ||
| Number of participants with available data | 69 (38%) | 120 (66%) | 88 (63%) | 79 (59%) | .. | ||
| Median | 24·8 (22·2–31·3) | 26·8 (23·0–30·4) | 27·1 (23·5–30·8) | 27·2 (24·1–32·1) | .. | ||
| Range | 18·4–51·2 | 17·4–45·9 | 14·3–46·7 | 19·8–46·3 | .. | ||
| Race | .. | .. | .. | .. | <0·0001* | ||
| Number of participants with available data | 168 (92%) | 160 (87%) | 134 (96%) | 125 (94%) | .. | ||
| Native American or Pacific Islander | 0 (0%) | 1 (1%) | 3 (2%) | 6 (5%) | .. | ||
| Asian | 1 (1%) | 5 (3%) | 2 (1%) | 9 (7%) | .. | ||
| Black or African American | 2 (1%) | 11 (6%) | 27 (19%) | 12 (9%) | .. | ||
| White | 165 (91%) | 143 (78%) | 102 (73%) | 98 (74%) | .. | ||
| Ethnicity | .. | .. | .. | .. | <0·0001* | ||
| Number of participants with available data | 176 (97%) | 171 (93%) | 138 (99%) | 128 (96%) | .. | ||
| Hispanic or Latino | 5 (3%) | 12 (7%) | 12 (9%) | 23 (17%) | .. | ||
| Not Hispanic or Latino | 171 (94%) | 159 (87%) | 126 (91%) | 105 (79%) | .. | ||
| Charlson comorbidity total score | .. | .. | .. | .. | <0·0001† | ||
| Number of participants with available data | 163 (90%) | 150 (82%) | 130 (94%) | 119 (89%) | .. | ||
| Median | 1·0 (0·0–1·0) | 0·0 (0·0–1·0) | 1·0 (0·0–2·0) | 1·0 (0·0–2·0) | .. | ||
| Range | 0·0–4·0 | 0·0–3·0 | 0·00–5·0 | 0·0–5·0 | .. | ||
| Charlson comorbidity index | |||||||
| Myocardial infarction | 3 (2%) | 0 (0%) | 2 (1%) | 2 (2%) | 0·45* | ||
| Congestive heart failure | 9 (5%) | 1 (1%) | 12 (9%) | 15 (11%) | 0·0060* | ||
| Peripheral vascular disease | 7 (4%) | 6 (3%) | 11 (8%) | 8 (6%) | 0·32* | ||
| Cerebrovascular disease | 8 (4%) | 2 (1%) | 7 (5%) | 8 (6%) | 0·16* | ||
| Dementia | 1 (1%) | 0 (0%) | 5 (4%) | 1 (1%) | 0·021* | ||
| Chronic pulmonary disease | 23 (13%) | 16 (9%) | 18 (13%) | 16 (12%) | 0·80* | ||
| Rheumatic disease | 7 (4%) | 4 (2%) | 4 (3%) | 3 (2%) | 0·81* | ||
| Peptic ulcer disease | 1 (1%) | 1 (1%) | 2 (1%) | 1 (1%) | 0·84* | ||
| Mild liver disease | 7 (4%) | 3 (2%) | 6 (4%) | 2 (2%) | 0·38* | ||
| Diabetes without chronic complications | 14 (8%) | 11 (6%) | 26 (19%) | 22 (17%) | 0·0012* | ||
| Diabetes with chronic complications | 4 (2%) | 2 (1%) | 9 (6%) | 9 (7%) | 0·020* | ||
| Hemiplegia or paraplegia | 0 (0%) | 0 (0%) | 3 (2%) | 0 (0%) | 0·018* | ||
| Renal disease | 12 (7%) | 6 (3%) | 19 (14%) | 19 (14%) | 0·0017* | ||
| Cancer | 38 (21%) | 11 (6%) | 19 (14%) | 20 (15%) | 0·0016* | ||
| Moderate or severe liver disease | 0 (0%) | 1 (1%) | 2 (1%) | 0 (0%) | 0·26* | ||
| Metastatic solid tumour | 6 (3%) | 0 (0%) | 4 (3%) | 1 (1%) | 0·068* | ||
| AIDS or HIV | 2 (1%) | 0 (0%) | 1 (1%) | 1 (1%) | 0·63* | ||
| APACHE II | .. | .. | .. | .. | <0·0001† | ||
| Number of participants with available data | 0 | 0 | 131 | 129 | .. | ||
| Median | .. | .. | 4·4 (3·8–6·7) | 7·6 (5·8–21·0) | .. | ||
| Range | .. | .. | [2·9–26·2] | [2·9–73·3] | .. | ||
| SARS-CoV-2 treatment | .. | .. | .. | .. | .. | ||
| Number of participants with available data | 0 | 0 | 131 | 129 | .. | ||
| IL-6 Inhibitor | .. | .. | 4 (3%) | 29 (22%) | <0·0001* | ||
| ACE inhibitor or ARB | .. | .. | 32 (23%) | 32 (24%) | 1·0* | ||
| Systemic corticosteroid | .. | .. | 69 (50%) | 98 (74%) | <0·0002* | ||
| Remdesivir | .. | .. | 63 (45%) | 82 (62%) | 0·017* | ||
| ECMO | .. | .. | 0 (0%) | 5 (4%) | 0·068* | ||
| Azithromycin | .. | .. | 55 (40%) | 83 (62%) | <0·0005* | ||
| Convalescent plasma | .. | .. | 10 (7%) | 6 (5%) | 0·46* | ||
Data are median (IQR), range, or n (%). APACHEII=Acute Physiology and Chronic Health Evaluation II. IL-6=interleukin-6. ACE=angiotensin-converting enzyme. ARB=angiotensin II receptor blockers. ECMO=extracorporeal membrane oxygenation.
χ2 test was applied to calculate p value.
F statistic was applied to calculate p value.
We used Olink Explore 1536 panel assay (Olink Proteomics [Uppsala, Sweden]), which uses proximity extension assay technology coupled to a readout methodology based on next generation sequencing (Illumina NovaSeq 6000, NextSeq 550, and NextSeq 2000; all manufactured by Illumina [San Diego, USA]; appendix 1 p 2), to quantify protein targets. Metabolon (Morrisville, NC, USA) did lipidomics using Sciex SelexION-5500 QTRAP and metabolomics using liquid chromatography-tandem mass spectrometry (LC-MS-MS; Thermo Fisher Scientific [Waltham, MA, USA]; appendix 1 pp 2–3). Pathway and network analysis was done using Ingenuity Pathway Analysis (Qiagen [Hilden, Germany]; appendix 1 p 3).
We developed a stacked ensemble model using a machine learning approach with AutoGluon-Tabular classifier (version 0.1.1b20210326). Input variables were preprocessed molecular markers, and data were stratified into train (80%) or test (20%) groups. 10-fold stratified cross-validation within our training set was used for model selection. The final model was evaluated on the test set. Additional details of data preprocessing and machine learning analysis are in appendix 1 (pp 3–4).
Proteomics and glycoproteomics using LC-MS-MS were done on 21 (89%) of 24 patients who had samples available from both before and after SAR-CoV-2 infection (appendix 1 pp 4–5). Publicly available single-cell RNA sequencing data were analysed from three COVID-19 studies using the in-house platform at nference (Cambridge, MA, USA; appendix 1 p 5).
Statistical analysis
To discover predictive markers of COVID-19 severity, we tested the statistical significance of individual molecules from the patients with severe and critical COVID-19 against those from outpatients, instead of uninfected controls (as this study focused on identifying biomarkers that would differentiate outpatients from the patients with more severe outcomes including hospital admission), using Student's t test. Adjusted p values were calculated by the Benjamini-Hochberg method. Additional details on the statistical analyses and model performances are in appendix 1 (p 5). We used a significance level of 0·05 to identify feature importance scores that performed well as predictors of COVID-19 severity outcomes.
Role of the funding source
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
Of the 455 patients who tested positive with COVID-19 between April 14, 2020, and Dec 1, 2020, 272 (60%) were admitted to hospital and 183 (40%) were outpatients (table 1). 182 age-matched and sex-matched uninfected individuals from the Mayo Clinic Biorepository were used as controls.12 Plasma samples from COVID-19 outpatients were obtained between 0 and 48 days (median 33 days [IQR 8–33]) after their first positive PCR test. For patients with severe COVID-19, plasma samples were collected between 0 and 18 days (median 2 days [0–5]), and for patients with critical COVID-19, between 0 and 47 days (median 3 days [1–7]). The median length of hospital stay was 5 days (IQR 3–7) for the patients with severe COVID-19 and 11 days for those with critical COVID-19 (IQR 8–25). A total of 1463 unique proteins, 902 lipids, and 1018 metabolites were analysed (appendix 1 pp 6, 8; appendix 2). Network analysis revealed enriched biological pathways and processes associated with severity and the results are summarised in appendix 1 (pp 6, 9–10, 17–18).
Using AutoGluon's tabular prediction algorithm, we generated a signature of 102 markers (53 proteins, 12 lipids, and 37 metabolites; table 2 ; appendix 1 p 15). Two baseline models were built for comparison; one using only IL-6 and the other using the cytokine storm panel of cytokines. A model built using all markers (protein, lipid, and metabolite markers) outperformed the baseline models in the held-out test dataset (ie, IL-6 and cytokine storm panel; p<0·0010), showing the superiority of the additional molecules profiled in this multiomics analysis over other established cytokines for predicting severity (appendix 1 pp 7, 16). A list of cytokine storm cytokines are shown in figure 1A and details on how the model can be used for new data is described in appendix 1 (p 6).
Table 2.
Predictive signature of COVID-19 severity
| Category | Importance score | |
|---|---|---|
| Linoleamide | Metabolite | 0·0793 |
| Leukotriene A4 hydrolase | Cytokine | 0·0500 |
| Oleamide | Metabolite | 0·0440 |
| Keratin, type I cytoskeletal 19 KRT19 | Cytokine | 0·0360 |
| C-C motif chemokine 7 | Cytokine | 0·0273 |
| Heme | Metabolite | 0·0227 |
| V-set and immunoglobulin domain-containing protein 4 VSIG4 | Cytokine | 0·0213 |
| 5-methyluridine | Metabolite | 0·0167 |
| Interleukin-15 | Cytokine | 0·0160 |
| Galectin-4 | Cytokine | 0·0133 |
| Lactate | Metabolite | 0·012 |
| Prostaglandin-H2 D-isomerase | Cytokine | 0·0093 |
| C-X-C motif chemokine 9 | Cytokine | 0·0087 |
| Integrin alpha-V | Cytokine | 0·008 |
| Leucylglycine | Metabolite | 0·008 |
| Odorant-binding protein 2b | Cytokine | 0·008 |
| Prolow-density lipoprotein receptor-related protein 1 | Cytokine | 0·0073 |
| 2,6-dihydroxy-benzoic acid | Metabolite | 0·0073 |
| Phosphatidylcholine (16:0/20:1) | Lipid | 0·0067 |
| Eicosanedioate | Metabolite | 0·006 |
| Docosadienoate | Metabolite | 0·006 |
| Bilirubin (E,Z or Z,E) | Metabolite | 0·0060 |
| Keratin, type I cytoskeletal 18 | Cytokine | 0·006 |
| γ-glutamylisoleucine | Metabolite | 0·006 |
| Dipeptidase 1 | Cytokine | 0·006 |
| Insulin-like growth factor-binding protein 6 | Cytokine | 0·006 |
| 4-aminophenol sulfate | Metabolite | 0·006 |
| Phosphatidylethanolamine (P-18:1/18:1) | Lipid | 0·0053 |
| Chymotrypsin-C | Cytokine | 0·0053 |
| Membrane primary amine oxidase | Cytokine | 0·0053 |
| γ-glutamylthreonine | Metabolite | 0·0053 |
| Integrin beta-6 | Cytokine | 0·0053 |
| Glucose | Metabolite | 0·0053 |
| Kallikrein-10 | Cytokine | 0·0053 |
| PE(O-16:0/18:1) | Lipid | 0·0047 |
| Acetoacetate | Metabolite | 0·0047 |
| Cathepsin B | Cytokine | 0·0047 |
| Diacylglycerol (18:1/18:2) | Lipid | 0·0047 |
| Ectonucleotide pyrophosphatase/phosphodiesterase family member 5 | Cytokine | 0·0047 |
| Carcinoembryonic antigen-related cell adhesion molecule 5 | Cytokine | 0·0047 |
| ICOS ligand | Cytokine | 0·0047 |
| 4-hydroxychlorothalonil | Metabolite | 0·0047 |
| Centrosomal protein of 85 kDa | Cytokine | 0·0047 |
| Serpin B5 | Cytokine | 0·0047 |
| T-cell surface glycoprotein CD1c | Cytokine | 0·0047 |
| Heneicosapentaenoate | Metabolite | 0·0047 |
| Glycoprotein hormones alpha chain | Cytokine | 0·0047 |
| Neurotrophin-4 | Cytokine | 0·0047 |
| Ras-related protein Rab-6A | Cytokine | 0·0047 |
| Epithelial cell adhesion molecule | Cytokine | 0·0040 |
| Dipeptidyl peptidase 2 | Cytokine | 0·0040 |
| Eicosapentaenoate | Metabolite | 0·0040 |
| Leukocyte immunoglobulin-like receptor subfamily A member 5 | Cytokine | 0·0040 |
| Calcineurin subunit B type 1 | Cytokine | 0·0040 |
| Androstenediol (3α, 17α) monosulfate | Metabolite | 0·0040 |
| Arachidonate | Metabolite | 0·0040 |
| δ-carboxyethyl hydroxychroman o | Metabolite | 0·0040 |
| Cortolone glucuronide | Metabolite | 0·0040 |
| Carnitine of C10H14O2 | Metabolite | 0·0040 |
| C-type lectin domain family 6 member A | Cytokine | 0·0040 |
| Theobromine | Metabolite | 0·0040 |
| Cysteine | Metabolite | 0·0040 |
| 2-hydroxydecanoate | Metabolite | 0·0033 |
| Lysophosphatidylethanolamine (18:1) | Lipid | 0·0033 |
| Triacylglycerol 55:7-fatty acid 20:3 | Lipid | 0·0033 |
| Integrin beta-1 | Cytokine | 0·0033 |
| PE(O-16:0/22:6) | Lipid | 0·0033 |
| Dual oxidase 2 | Cytokine | 0·0033 |
| Retbindin | Cytokine | 0·0033 |
| Alpha-L-iduronidase | Cytokine | 0·0033 |
| N-methylpipecolate | Metabolite | 0·0033 |
| 4-hydroxyphenylpyruvate | Metabolite | 0·0033 |
| Hemoglobin subunit theta-1 | Cytokine | 0·0033 |
| Phosphatidylinositol (18:0/20:3) | Lipid | 0·0033 |
| Phenylacetylcarnitine | Metabolite | 0·0033 |
| Alpha-amylase 2B | Cytokine | 0·0033 |
| Cadherin-6 | Cytokine | 0·0033 |
| Ursodeoxycholate | Metabolite | 0·0033 |
| Phospholipase A2 group XV | Cytokine | 0·0027 |
| Protein GOLM2 | Cytokine | 0·0027 |
| Pentadecanoate | Metabolite | 0·0027 |
| Retinoic acid receptor responder protein 2 | Cytokine | 0·0027 |
| Angiopoietin-2 | Cytokine | 0·0027 |
| C-X-C motif chemokine 11 | Cytokine | 0·0027 |
| Pancreatic alpha-amylase | Cytokine | 0·0027 |
| Carbonic anhydrase 1 | Cytokine | 0·0027 |
| Neuropilin-1 | Cytokine | 0·002 |
| Inositol 1,4,5-triphosphate receptor associated 2 | Cytokine | 0·002 |
| Nucleoside diphosphate kinase 3 | Cytokine | 0·002 |
| Arabinose | Metabolite | 0·002 |
| Diacylglycerol (16:1/20:4) | Lipid | 0·002 |
| 3-methyl catechol sulfate | Metabolite | 0·002 |
| TBC1 domain family member 5 | Cytokine | 0·002 |
| TAG56:6-FA18:3 | Lipid | 0·002 |
| Dihydroceramide (18:1) | Lipid | 0·002 |
| Sebacate | Metabolite | 0·002 |
| EF-hand calcium-binding domain-containing protein 4B | Cytokine | 0·002 |
| Hydroxyacylglutathione hydrolase, mitochondrial | Cytokine | 0·002 |
| Cysteinyl-glycine, oxidize | Metabolite | 0·002 |
| Trizma acetate | Metabolite | 0·002 |
| Phosphatidylethanolamine (16:0/20:4) | Lipid | 0·002 |
| Cysteinylglycine disulfide | Metabolite | 0·002 |
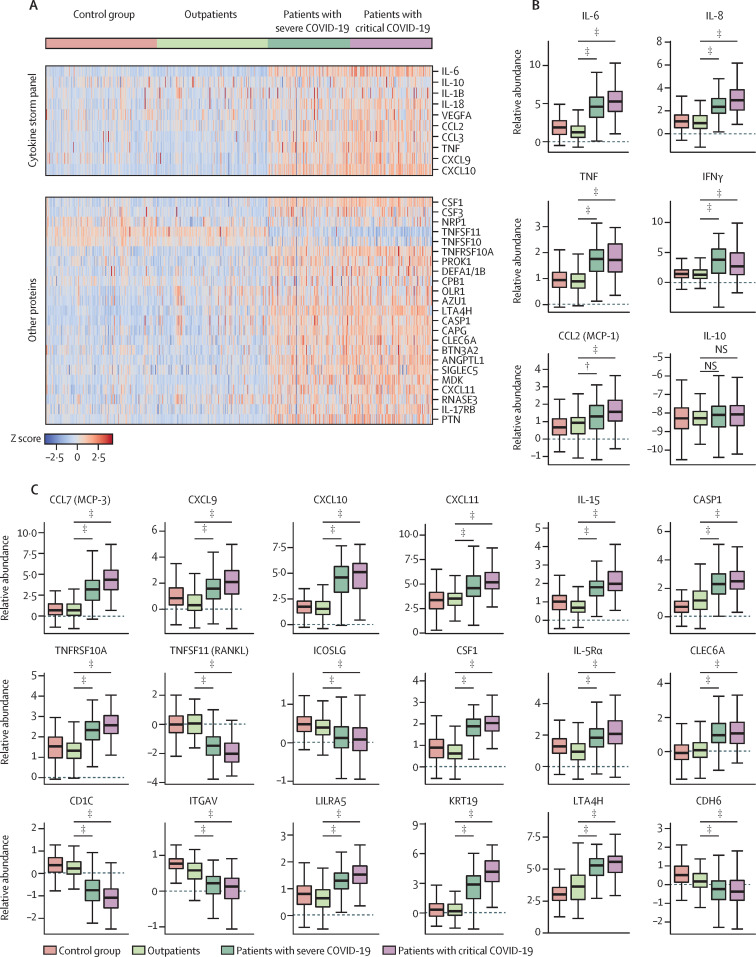
Figure 1.
Relative levels of known cytokine storm panel and other proteins in plasma among the study participants (A). Distribution of plasma levels (relative abundance; log2 values) across the patient subgroups for select cytokine storm cytokines (B) and other protein markers (C).
IL=interleukin. VEGFA=vascular endothelial growth factor A. CCL=C-C motif chemokine. TNF=tumour necrosis factor. CXCL=C-X-C motif chemokines. CSF1=macrophage colony-stimulating factor 1. NRP1=neuropilin-1. TNFSF=tumour necrosis factor ligand superfamily member. PROK1=prokineticin-1. DEFA1=neutrophil defensin 1. CPB1=carboxypeptidase B. OLR1=oxidized low-density lipoprotein receptor 1. AZU1=azurocidin. LTA4H=leukotriene A-4 hydrolase. CASP1=caspase-1. CAPG=macrophage-capping protein. CLEC6A=c-type lectin domain family 6 member. BTN3A2=butyrophilin subfamily 3 member A2. ANGPTL1=angiopoietin-related protein 1. NS=no significance. SIGLEC5=sialic acid-binding Ig-like lectin 5. MDK=midkine. RNASE3=eosinophil cationic protein. PTN=pleiotrophin. TNFRSF=tumour necrosis factor receptor superfamily member. ICOSLG=ICOS ligand. CD1C=t-cell surface glycoprotein CD1c. ITGAV=integrin alpha-V. LILRA5=leukocyte immunoglobulin-like receptor subfamily A member 5. KRT19=keratin, type I cytoskeletal 19. CDH6=cadherin-6. *Adjusted p≤0·05. †Adjusted p≤0·01. ‡Adjusted p≤0·001.
Machine learning and individual statistical analysis identified several proteins that were significantly different between the severity groups. Cytokines associated with cytokine storm and macrophage activation syndromes showed an expected association with disease severity; concentrations of IL-6, IL-8, C-C motif chemokine 2 (CCL2), vascular endothelial growth factor A (VEGFA), TNF, and interferon (IFN)-γ were positively associated with disease severity (figure 1A, 1B). IL-10 and IL-1β did not show such an association. Several other cytokines and chemokines were positively associated with severity, including C-C motif chemokine 7 (CCL7), C-X-C motif chemokines 9, 10 and 11 (CXCL9, CXCL10, CXCL11), IL-15, azurocidin (AZU1) and midkine (MDK; figure 1A, 1C). Markers of apoptosis, including caspase-1 (CASP1) and TNF receptor superfamily member 10A (TNFRSF10A), were higher in patients with greater severity of disease (figure 1C). There were decreased concentrations of TNF ligand superfamily member 10 (TNFSF10), TNF ligand superfamily member 11 (TNFSF11), and ICOS ligand (ICOSLG) in patients with greater severity of disease (figure 1A, 1C). Increased concentrations of apoptotic marker prolow-density lipoprotein receptor-related protein 1 (LRP1) and V-set and immunoglobulin domain-containing protein 4 (VSIG4), which are involved in the complement pathway, were both associated with greater severity (appendix 1 p 11). The concentrations of haematopoietic mediators including macrophage colony-stimulating factor (CSF1), and interleukin-5 receptor subunit alpha (IL-5Rα) were also higher in patients with greater disease severity (figure 1C). Concentrations of glycan-binding molecules C-type lectin domain family 6 member A (CLEC6A), sialic acid-binding Ig-like lectin 5 (SIGLEC5), and galectin-9 (LGALS9) were positively associated with severity, and N-acetylgalactosaminyltransferase 7 (GALNT7) were negatively associated with severity (figure 1A, 1C; appendix 1 p 11). Increased concentrations of heparan sulfate glucosamine 3-O-sulfotransferase 3B1 (HS3ST3B1) and syndecan-1 (SDC1), both related to proteoglycans, were associated with greater severity (appendix 1 p 11). Among cell surface receptors, T-cell surface glycoprotein CD1c (CD1C) and integrin alpha-V (ITGAV) were negatively associated with severity, and leukocyte immunoglobulin-like receptor subfamily A member 5 (LILRA5), lymphocyte activation gene 3 protein (LAG3) and macrophage-capping protein (CAPG) were positively associated (figure 1A, 1C; appendix 1 p 11). Increased levels of keratin, type I cytoskeletal 19 (KRT19) was associated with greater severity, but cartilage oligomeric matrix protein (COMP) and cartilage acidic protein 1 (CRTAC1) were downregulated with increasing disease severity (figure 1C; appendix 1 p 11). Concentrations of leukotriene A-4 hydrolase (LTA4H), and matrilysin (MMP7) were higher in the severe and critical cohorts than in outpatients, and kallikrein-13 (KLK13) was lower in the severe and critical cohorts than in outpatients. Prokineticin-1 (PROK1), angiopoietin-related protein 1 (ANGPTL1), and ephrin type-B receptor 4 (EPHB4), which are associated with vascular remodelling, were positively associated (figure 1A; appendix 1 p 11). Calcitonin (CALCA) and SPARC-related modular calcium-binding protein 1 (SMOC1) were higher in the severe and critical cohorts than in outpatients, whereas cadherin-6 (CDH6) and neuropeptide Y (NPY) were lower in the severe and critical cohorts than in outpatients (figure 1C; appendix 1 p 11).
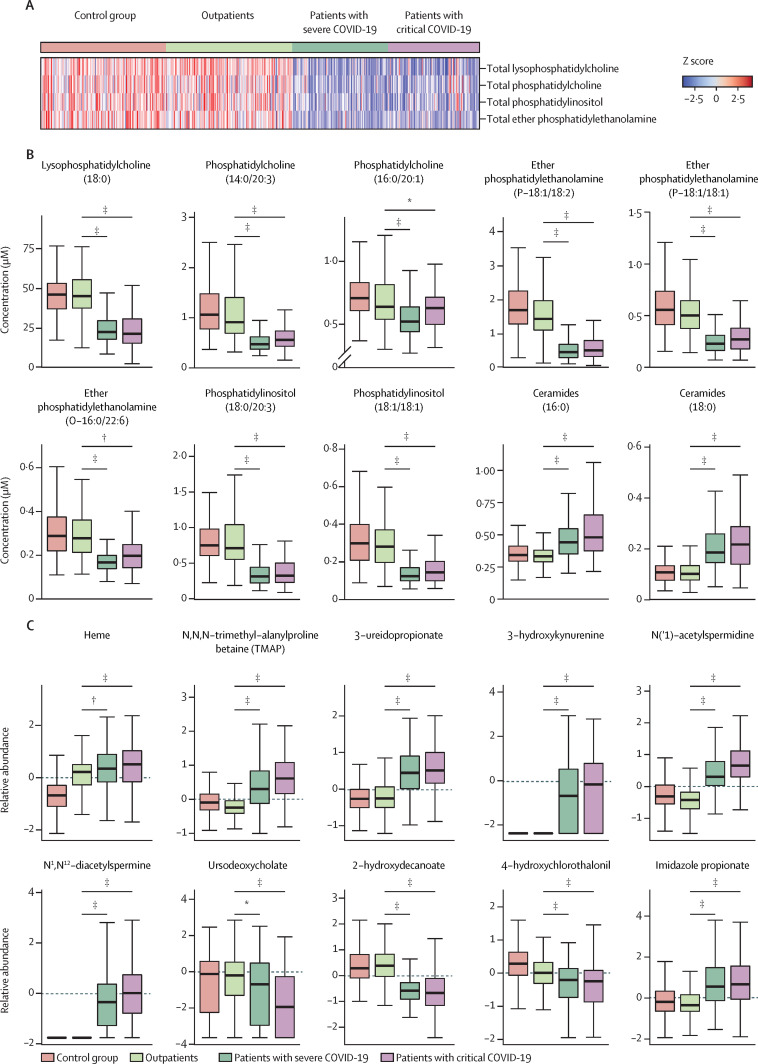
An overview of the relative abundance of 14 classes of lipids across disease severity categories is shown in appendix 1 (p 12). Although the abundance of classes such as diglycerides remained unchanged across the groups, lysophosphatidylcholine, phosphatidylcholine, ether phosphatidylethanolamine, and phosphatidylinositol were lower in the hospitalised patients than in the outpatients (figure 2A ). Among the 102 predictive biomarkers of COVID-19 severity, 12 were lipids. Individual statistical analysis identified several species of phospholipids that were attenuated in hospitalised participants including lysophosphatidylcholine (18:0), phosphatidylcholine (14:0/20:3), phosphatidylcholine (16:0/20:1), ether phosphatidylethanolamine (P-18:1/18:2), ether phosphatidylethanolamine (P-18:1/18:1), ether phosphatidylethanolamine (O-16:0/22:6), phosphatidylinositol (18:0/20:3), and phosphatidylinositol (18:1/18:1). In contrast to a general decrease in phospholipids with increasing levels of disease severity, concentrations of select species of ceramides, including ceramides (16:0) and ceramides (18:0), were increased in hospitalised participants (figure 2B).
Figure 2.
(A) Heatmap of selective classes of phospholipids showing decreased changes in severe and critical cases of COVID-19 in which each rectangle represents a participant. Distribution of (B) lipids and (C) metabolites showing significant changes across participant groups
NS=no significance. *Adjusted p≤0·05. †Adjusted p≤0·01. ‡Adjusted p≤0·001.
Individual statistical analysis uncovered metabolites that showed significant elevation in hospitalised patients compared with outpatients including heme, N,N,N-trimethyl-alanylproline betaine (TMAP), 3-ureidopropionate, 3-hydroxykynurenine, and polyamine metabolites N(‘1)-acetylspermidine and N1,N12-diacetylspermine. Several metabolites related to the host gut microbiome were also significantly associated with disease severity; hospitalised patients were associated with decreased concentrations of ursodeoxycholate and 2-hydroxyodecanoate (a medium-chain fatty acid), and increased concentrations of a histidine-derived metabolite, imidazole propionate (figure 2C).
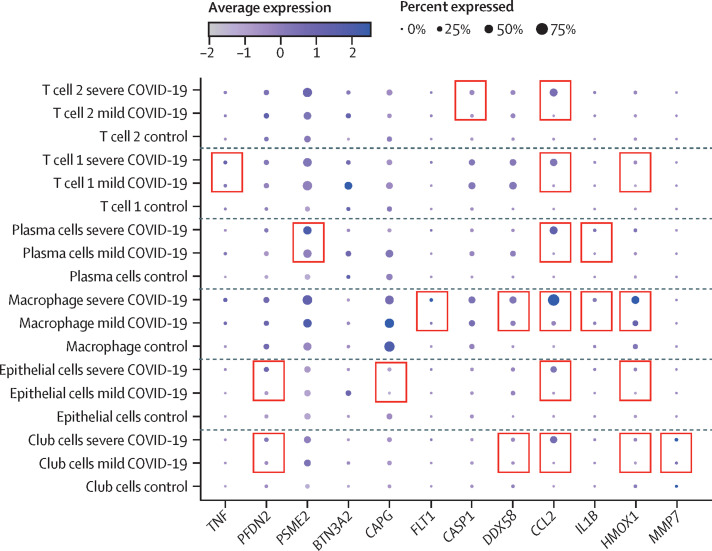
To analyse cellular mRNA expression of predictive cytokine markers, we identified three publicly-available studies in which cells isolated from patients with COVID-19, who had disease ranging from mild to severe, were profiled by RNA sequencing.13, 14, 15 We observed a similar upregulation on the basis of single-cell RNA-sequencing data in the severe group for 17 proteins that were identified from our cytokine analysis (figure 3 ; appendix 1 pp 12–13). Details of upregulated genes are in appendix 1 (p 6).
Figure 3.
Significantly upregulated genes in patients with severe COVID-19 compared with patients with mild COVID-19 and control participants in both a single-cell RNA sequencing study15 and the current study
The pink boxes denote genes that are upregulated in the severe cohort as compared with the mild cohort in both stem-cell RNA sequencing and plasma proteomics analysis. T cell 1, T cell 2, plasma cells, macrophages, epithelial cells, and club cells represent the various clusters of cells described in the single-cell RNA sequencing study. TNF=tumour necrosis factor. PFDN2=prefoldin subunit 2. PSME2=proteasome activator complex subunit 2. BTN3A2=butyrophilin subfamily 3 member A2. CAPG=macrophage-capping protein. FLT1=vascular endothelial growth factor receptor 1. CASP1=caspase-1. DDX58=antiviral innate immune response receptor RIG-I. CCL2=C-C motif chemokine 2. IL1B=antiviral innate immune response receptor RIG-I. HMOX1=heme oxygenase 1. MMP7=matrilysin.
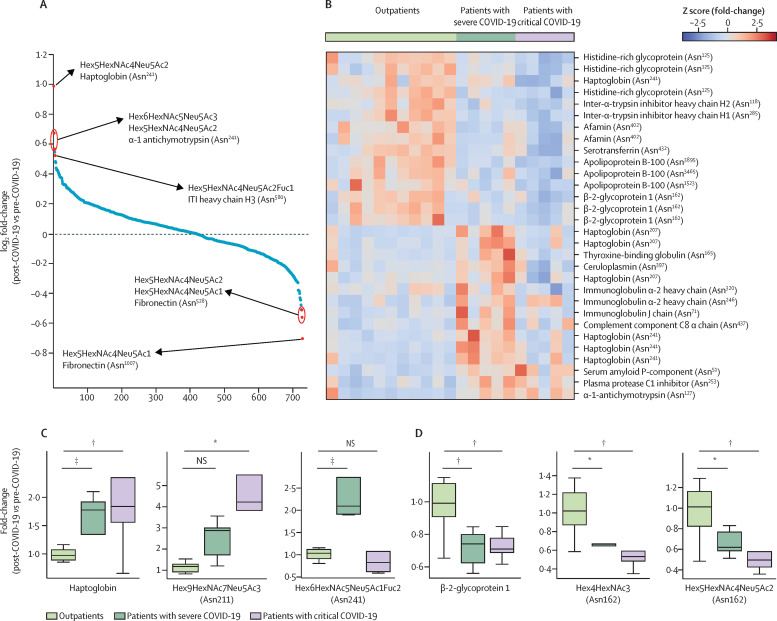
Details of the 21 patients whose pre-COVID-19 plasma samples were available and used for proteomics and glycoproteomics analysis are in appendix 1 (p 14). 472 proteins and 732 glycopeptides were quantified in all 21 patients (figure 4A ). The abundance values of the glycopeptides were used to calculate the ratio of post-COVID-19 glycopeptides to pre-COVID-19 glycopeptides for each patient. These ratios were then compared between the three groups (outpatients, patients with severe COVID-19, and patients with critical COVID-19) and one-way ANOVA was done, which identified 114 glycopeptides (derived from 62 glycosylation sites of 47 glycoproteins; p<0·05) that were significantly different between the groups (figure 4B). Glycosylation was found to be decreased on Asn1523, Asn3465, and Asn3595 of apolipoprotein B-100 and on Asn402 of afamin (figure 4B). Overall, protein concentrations of these molecules were reduced in post-COVID-19 samples when compared with pre-COVID-19 samples in patients with severe and critical disease but not in outpatients. Additionally, one glycopeptide of haptoglobin (Asn211) with a composition of Hex9HexNAc7NeuAc3 was not altered in outpatients but elevated in patients with severe disease and in patients with critical disease (figure 4C). The distribution of total haptoglobin and its glycopeptides (Asn207 and Asn241) is shown in figure 4C and appendix 1 (p 14). Sialylated complex-type glycan containing glycopeptides derived from β-2-glycoprotein 1 (APOH; figure 4D) and kininogen-1 (appendix 1 p 14) were significantly decreased in patients with critical COVID-19 and kininogen-1 Asn72 was increased (appendix 1 p 14). Protein concentrations of α-1-acid glycoprotein 1 (ORM1) were significantly elevated in patients who were admitted to hospital.
Figure 4.
LC-MS-MS-based glycoproteomics comparing plasma glycopeptides in patients before and after they contracted COVID-19, (A) waterfall plot showing variation in median fold change of each glycopeptide on log2 scale from all patients, (B) heatmap of 34 glycopeptides (p≤0·050) in patients, (C) box plots showing variation in two representative glycopeptides of haptoglobin and β-2-glycoprotein 1 and (D) variation in two representative glycopeptides of haptoglobin and β-2-glycoprotein 1 across different patient groups
B is colour coded for WHO OSCI scores; protein names with site of glycosylation are given for each glycopeptide and identical names refer to different glycan chains at the same site of glycosylation. LC-MS-MS=liquid chromatography-tandem mass spectrometry. Hex=Hexose. NAc=N-acetylhexosamine. Neu5Ac=N-acetylneuraminic acid. Fuc=Fucose. OSCI=ordinal scale of clinical improvement. *Adjusted p≤0·05. †Adjusted p≤0·01. ‡Adjusted p≤0·001.
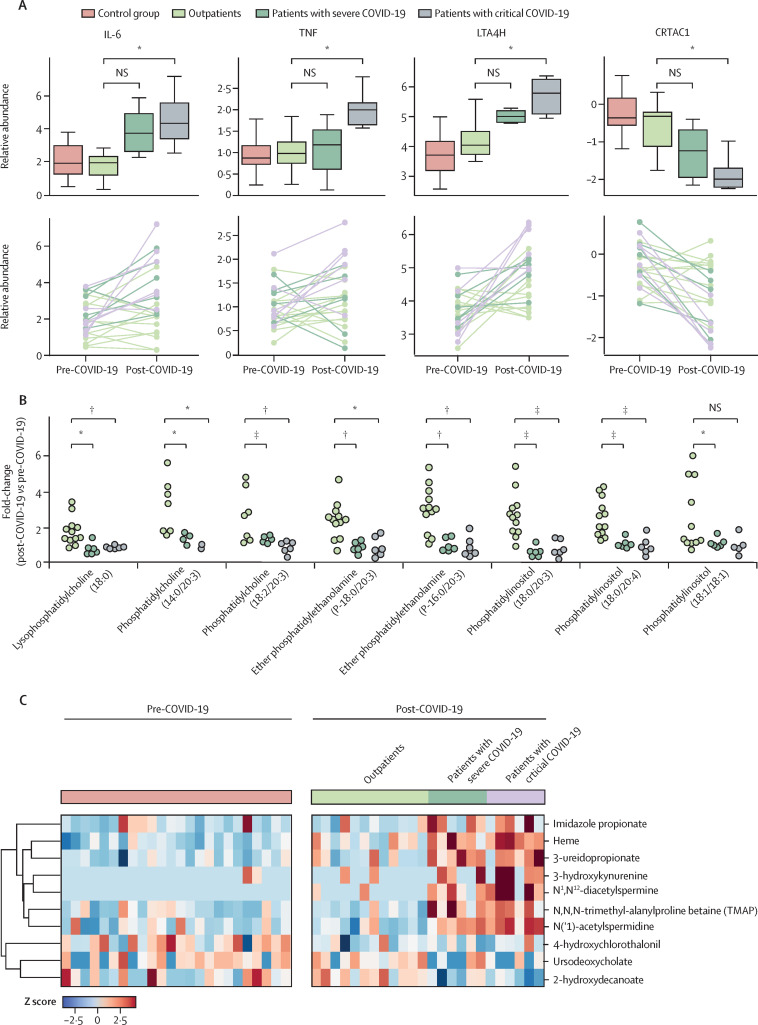
We used 24 paired samples to do an expanded multiomics profiling and this paired analysis allowed us to examine how various molecules were altered in these individuals following infection. Notably, most molecules followed the patterns observed in the design described above, including IL-6, TNF, and LTA4H (ie, in patients with severe and critical COVID-19, these molecules were found in higher concentrations in the post-COVID-19 cohorts than in the pre-COVID-19 cohorts; figure 5A ). Following COVID-19, CRTAC1 was significantly decreased in patients with critical disease, and non-significantly decreased in patients with severe disease. Reduced concentrations of lysophosphatidylcholine, phosphatidylcholine, phosphatidylinositol, and ether phosphatidylethanolamine species were observed in hospitalised patients following COVID-19, supporting the association of these lipids with severe and critical COVID-19 outcomes (figure 5B). Metabolites including heme, ursodeoxycholate, 3-ureidopropionate, and TMAP across the matched samples also showed a similar trend of changes as observed in the cross-sectional analysis (figure 5C).
Figure 5.
Distribution of cytokine (A), lipid (B), and metabolite markers (C) in a specialised subset of patients with COVID-19 with matched pre-COVID-19 plasma samples.
IL=interleukin. TNF=tumour necrosis factor. LTA4H=leukotriene A-4 hydrolase. CRTAC1=cartilage acidic protein 1. NS=no significance. *Adjusted p≤0·05. †Adjusted p≤0·01. ‡Adjusted p≤0·001.
Discussion
To our knowledge, this study of 637 individuals, which included 455 patients with COVID-19 at different levels of severity, represents the largest profiling of COVID-19 plasma samples using an integrative multiomics approach to date, using an integrative multiomics approach. Additionally, we present data from a unique collection of 24 patients with COVID-19 for whom matched pre-COVID-19 plasma samples were available. We report predictive biomarkers of severity in COVID-19. We saw that several molecules associated with the cytokine storm syndrome were associated with severity, as expected, including IL-6.10, 11 Other markers of systemic immune response, including CSF1, IL-5Rα, and IL-15 showed a similar association. We also identified additional plasma protein markers that have been implicated in inflammation, apoptosis, and other important cellular processes, including several that have not previously been described in the context of COVID-19. SIGLEC5, which has been hypothesised to be involved in the ligation of SARS-CoV-2 glycoprotein with CD33, was elevated in patients with severe and critical COVID-19.16 The elevated concentrations of CLEC6A, CCL7, AZU1, CAPG, and LILRA5 in these hospitalised patients are suggestive of the innate immune response associated with severity. Elevated CXCL9, CXCL10, and CXCL11, which are secreted by chemotactically activated immune cells, are indicative of strong interferon-mediated responses.17 LAG3 and LGALS9, involved in lymphocyte function, were elevated in the severe and critical groups; ICOSLG, however, was reduced. Although proinflammatory enzyme LTA4H was high in patients with severe and critical disease, VSIG4, a phagocytic receptor with anti-inflammatory effects, also showed a positive association with severity; this is a novel association that we report.18 Patients with severe and critical outcomes showed elevated concentrations of apoptotic markers TNFRSF10A and FAS, but concentrations of TNFSF10 and TNTFSF11 were reduced in these groups. Interestingly, COMP, a known biomarker of osteoarthritis and suppressor of apoptosis, was downregulated in patients with severe and critical disease.
SDC1, a proteoglycan, known to be involved in COVID-19 pathogenesis, was elevated in patients with severe and critical COVID-19. MDK, a cytokine and growth factor, was also elevated in these groups, along with one of its interactors, LRP1. CD1C, a T-cell surface glycoprotein, was negatively associated with severity; this is consistent with a 2020 report that CD1C+ T cells preferentially migrate from blood to lungs in patients with severe COVID-19.19 MMP7, which has been reported to be elevated in patients with post-acute sequelae of SARS-CoV-2 infection who previously needed admission to intensive care, was elevated in patients with severe and critical COVID-19.20 CRTAC1 was negatively associated with severity; this might be indicative of the poor health status of alveolar type-2 epithelial cells in patients with severe COVID-19 as has been previously described in the context of interstitial lung disease.11, 21 Angiogenesis-related proteins were elevated in patients with severe and critical COVID-19, including VEGFA and ANGPTL1, along with EPHB4 and dimethylarginase-1 (DDAH1; a promoter of nitric oxide biosynthesis and angiopoiesis); this is in keeping with previous studies that describe vascular angiogenesis as an important feature of COVID-19.22 Consistent with previous reports of consumption of components of the coagulation system,23 KLK13 concentrations were downregulated in patients with severe and critical COVID-19.
We found differences in the concentrations of several lipid classes between patients with severe and critical COVID-19 and outpatients with COVID-19. In contrast to our finding of decreased concentrations of lysophosphatidylcholine in hospitalised patients, previous studies have reported increased lysophosphatidylcholine in patients with COVID-19.7, 8 Although lysophosphatidylcholine has been reported as a proinflammatory mediator in the past, a 2020 study questioned whether lysophosphatidylcholine exerts anti-inflammatory and vasoprotective roles.24 Also, decreased lysophosphatidylcholine has been reported in patients with pulmonary arterial hypertension.24 Most importantly, lysophosphatidylcholine inhibits the release of IL-6; thus, decreased lysophosphatidylcholine in hospitalised patients corresponds with increased IL-6 in these patients.25 In addition to reduction of lysophosphatidylcholine, phosphatidylcholine was also found to be decreased in hospitalised patients, consistent with other studies.7, 8 Plasmalogen ether phosphatidylethanolamine is known to possess antioxidant properties and its reduction in hospitalised patients suggests increased oxidative stress.26 Phosphatidylinositol acts as an antiviral mediator and it has been reported to attenuate infection in respiratory syncytial virus.27 As an inhibitor of IL-8, phosphatidylinositol suppresses infection and viral propagation.27 Decreased phosphatidylinositol is in line with elevated IL-8 in patients with SARS-CoV-2. Reduced ether phosphatidylethanolamine and phosphatidylinositol could be suggestive of induced oxidative stress and poorly controlled viral propagation in hospitalised patients. Select species of ceramides were increased in hospitalised patients; elevated ceramide is related to pulmonary cell apoptosis and emphysema-like disease and reduced ceramide protects against oxidative stress.28 Notably, increased serum ceramides have been described in patients with severe SARS-CoV-2.7
Several metabolite predictors of COVID-19 severity were also discovered. High concentrations of heme in patients admitted to hospital is consistent with reports that SARS-CoV-2 causes elevation of free heme.29 TMAP, a marker of reduced kidney function, showed a positive association with increasing severity, which supports findings from a 2021 study that reported the same trend in patients with severe disease.30 Polyamines facilitate virus replication by aiding cellular attachment; the increased polyamine concentrations in hospitalised patients might indicate increased disease severity.31 We discovered accumulation of 3-ureidopropionate, a metabolite in pyrimidine metabolism, in the severe and critical cohorts. 3-ureidopropionate increases reactive oxygen species and inhibits complex V.32 Anosmia has been reported by patients with COVID-19 and 3-ureidopropionate is a precursor of β alanine, which is required for the synthesis of carnosine that protects the olfactory sensory neurons and sustentacular cells.32 Olfactory sensory neurons are damaged during infection with SARS-CoV-2, causing anosmia; thus, we speculate that inhibition of complex V by 3-ureidopropionate might be related to damaged neurons and anosmia. Our results reflected the alteration of the gut microbiome in patients admitted to hospital with COVID-19. 2-hydroxydecanoate exerts anti-viral properties and has been known to inhibit the viral replications;33 its decrease in hospitalised patients corresponds with severe infection. Antioxidant ursodeoxycholate, an important bile acid in SARS-CoV-2 infection due to its antioxidant and antiapoptotic properties that can inhibit proinflammatory cytokine storm, was decreased with increasing disease severity, supporting its negative association with cytokine storm.34
APOH, known to drive hepatitis virus retention by direct interaction and induce endoplasmic reticulum stress, was reduced in patients with severe and critical disease for whom paired pre-COVID-19 samples were analysed.35 Kininogen-1, which plays an important role in prekallikrein-kinin axis, was also reduced. ORM1, however, was increased; this is the first demonstration in the context of COVID-19. The glycosylation levels of APOB were reduced in hospitalised patients; interestingly, N-glycosylation mutant APOB in liver has been known to lead to endoplasmic reticulum stress and insulin resistance, suggesting glycosylation of APOB is important for its sustained function.36
Our study revealed novel molecules including cytokines, lipids, and metabolites as predictive biomarkers of severe and critical outcomes after COVID-19 infection. Importantly, these markers were found to be altered early in the course of the disease, before or around the time of hospital admission, suggesting a close link between inflammation and adverse outcomes. The markers described in this study improved the accuracy of prediction of COVID-19 severity over the classic cytokine storm panel of cytokines. For example, although previous studies have suggested IL-6 and TNF concentrations in plasma as predictors of COVID-19 severity, our study identified a multiomic signature of 102 analytes that perform better than them.
Even though this is a retrospective study, the samples were drawn at the baseline when the patients had typically not yet reached that worst outcome. Although we discovered potentially predictive markers, they do not necessarily imply causation. The number of biomarkers collected was greater than the size of our cohort, which can lead to concerns of overfitting. However, the sophisticated AutoGluon modelling approach avoided overfitting as evidenced by our high accuracy in the held-out test set. Without an external dataset, our study could not evaluate whether these models would extend to cohorts that are not well represented by the Mayo Clinic population and this remains a major limitation of the study. We are also limited by the changing nature of treatments and the disease itself over the course of the pandemic, offering a group of confounders that cannot be remedied in a retrospective design.
Data sharing
All clinical and multiomics data of deidentified subjects are available in the published appendices; personal health information is protected, as required by law. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD029376 under the project name “Large-scale multiomics analysis identifies novel early markers of COVID-19 severity”; the repository will make the dataset publicly available upon publication and it will be accessible with the dataset identifier. A code snippet that shows the usage of AutoGluon is provided in appendix 1.
Declaration of interests
PK-M, TH, and AJV are employees of nference. JCO receives grants from nference and personal fees from Elsevier and Bates College, outside the submitted work. ADB is supported by grants from NIAID (AI110173 and AI120698), Amfar (109593), and Mayo Clinic (HH Shieck Khalifa Bib Zayed Al-Nahyan Named Professorship of Infectious Diseases). ADB is a paid consultant for AbbVie, Gilead, Freedom Tunnel, Pinetree Therapeutics, Primmune, Immunome, MarPam, Rion, and Flambeau Diagnostics; is a paid member of the DSMB for Corvus Pharmaceuticals, Equilium, and Excision Biotherapeutics; has received fees for speaking for Reach MD, Peer Voice, and Medscape; owns equity for scientific advisory work in Zentalis Rion and nference; and is founder and President of Splissen Therapeutics.
Acknowledgments
Acknowledgments
Funding for the molecular profiling was supported by the generosity of Eric and Wendy Schmidt. We are grateful to Jessica L Lesko for coordinating the project. We thank Jon Harrington, Josh Bublitz, and Terra Lasho for organising and sharing patient and clinical data. We acknowledge Katelyn A Reed for assistance with institutional review boards and logistical support. We would also like to thank Dong-Gi Mun for help with samples.
Contributors
SKB, AKM, KG, JRW, GJ, JCO, ADB, and AP had access to all data in the study and all other authors had access to the datasets they were involved in but not to all data due to patient confidentiality agreements. All authors had final responsibility for the decision to submit for publication. SKB, AKM, MMP, NC, SA-K, RC, MES, BRK, SKGG, RJS, AASA, ADB, and AP conceived and designed the study. SKB, AKM, KG, MGR, and MS analysed or interpreted data. KG, MS, and RS did the proteomics and glycoproteomics experiments. MMP, SA-K, AA-S, AASA, JDY, BSP, JEO, JCO, and ADB provided samples and clinical data. JRW, AKM, and GJ did the machine learning and statistical analysis. AKM and RMZ developed visual representation of omics data. SKB, AKM, KG, MGR, MS, PK-M, TH, AJV, JRW, GJ, JCO, ADB, and AP wrote the manuscript. PK-M, TH, and AJV analysed and interpreted single-cell RNA sequencing data. SD did the pathway analysis. NC, RC, MES, SD, BRK, SKGG, and RJS critically reviewed the manuscript. SKB and AKM contributed equally as co-first authors. All authors critically read and approved the manuscript.
Supplementary Materials
References
- 1.Rosas IO, Bräu N, Waters M, et al. Tocilizumab in hospitalized patients with severe COVID-19 pneumonia. N Engl J Med. 2021;384:1503–1516. doi: 10.1056/NEJMoa2028700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horby P, Lim WS, Emberson JR, et al. Dexamethasone in hospitalized patients with COVID-19. N Engl J Med. 2021;384:693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Group RC. Tocilizumab in patients admitted to hospital with COVID-19 (RECOVERY): a randomised, controlled, open-label, platform trial. Lancet. 2021;397:1637–1645. doi: 10.1016/S0140-6736(21)00676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavalli G, De Luca G, Campochiaro C, et al. Interleukin-1 blockade with high-dose anakinra in patients with COVID-19, acute respiratory distress syndrome, and hyperinflammation: a retrospective cohort study. Lancet Rheumatol. 2020;2:e325–e331. doi: 10.1016/S2665-9913(20)30127-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feldmann M, Maini RN, Woody JN, et al. Trials of anti-tumour necrosis factor therapy for COVID-19 are urgently needed. Lancet. 2020;395:1407–1409. doi: 10.1016/S0140-6736(20)30858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel J, Beishuizen A, Ruiz XB, et al. A randomized trial of otilimab in severe COVID-19 pneumonia (OSCAR) medRxiv. 2021 doi: 10.1101/2021.04.14.21255475. published online April 17. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song JW, Lam SM, Fan X, et al. Omics-driven systems interrogation of metabolic dysregulation in COVID-19 pathogenesis. Cell Metab. 2020;32:188–202. doi: 10.1016/j.cmet.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barberis E, Timo S, Amede E, et al. Large-scale plasma analysis revealed new mechanisms and molecules associated with the host response to SARS-CoV-2. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21228623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas T, Stefanoni D, Reisz JA, et al. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight. 2020;5 doi: 10.1172/jci.insight.140327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Su Y, Chen D, Yuan D, et al. Multi-omics resolves a sharp disease-state shift between mild and moderate COVID-19. Cell. 2020;183:1479–1495. doi: 10.1016/j.cell.2020.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Overmyer KA, Shishkova E, Miller IJ, et al. Large-scale multi-omic analysis of COVID-19 severity. Cell Syst. 2021;12:23–40. doi: 10.1016/j.cels.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olson JE, Ryu E, Hathcock MA, et al. Characteristics and utilisation of the Mayo Clinic Biobank, a clinic-based prospective collection in the USA: cohort profile. BMJ Open. 2019;9 doi: 10.1136/bmjopen-2019-032707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chua RL, Lukassen S, Trump S, et al. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat Biotechnol. 2020;38:970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- 14.Wilk AJ, Rustagi A, Zhao NQ, et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bost P, Giladi A, Liu Y, et al. Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell. 2020;181:1475–1488. doi: 10.1016/j.cell.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murch SH. Common determinants of severe COVID-19 infection are explicable by SARS-CoV-2 secreted glycoprotein interaction with the CD33-related Siglecs, Siglec-3 and Siglec-5/14. Med Hypotheses. 2020;144 doi: 10.1016/j.mehy.2020.110168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vanderboom PM, Mun DG, Madugundu AK, et al. Proteomic signature of host response to SARS-CoV-2 infection in the nasopharynx. Mol Cell Proteomics. 2021;20 doi: 10.1016/j.mcpro.2021.100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Diao B, Guo S, et al. VSIG4 inhibits proinflammatory macrophage activation by reprogramming mitochondrial pyruvate metabolism. Nat Commun. 2017;8 doi: 10.1038/s41467-017-01327-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sánchez-Cerrillo I, Landete P, Aldave B, et al. COVID-19 severity associates with pulmonary redistribution of CD1c+ DCs and inflammatory transitional and nonclassical monocytes. J Clin Invest. 2020;130:6290–6300. doi: 10.1172/JCI140335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chun HJ, Coutavas E, Pine AB, et al. Immunofibrotic drivers of impaired lung function in postacute sequelae of SARS-CoV-2 infection. JCI Insight. 2021;6 doi: 10.1172/jci.insight.148476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayr CH, Simon LM, Leuschner G, et al. Integrative analysis of cell state changes in lung fibrosis with peripheral protein biomarkers. EMBO Mol Med. 2021;13 doi: 10.15252/emmm.202012871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ackermann M, Verleden SE, Kuehnel M, et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in COVID-19. N Engl J Med. 2020;383:120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lipcsey M, Persson B, Eriksson O, et al. The outcome of critically ill COVID-19 patients is linked to thromboinflammation dominated by the kallikrein/kinin system. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.627579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen C, Luo F, Wu P, et al. Metabolomics reveals metabolite changes of patients with pulmonary arterial hypertension in China. J Cell Mol Med. 2020;24:2484–2496. doi: 10.1111/jcmm.14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rolin J, Vego H, Maghazachi AA. Oxidized lipids and lysophosphatidylcholine induce the chemotaxis, up-regulate the expression of CCR9 and CXCR4 and abrogate the release of IL-6 in human monocytes. Toxins (Basel) 2014;6:2840–2856. doi: 10.3390/toxins6092840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wallner S, Orsó E, Grandl M, Konovalova T, Liebisch G, Schmitz G. Phosphatidylcholine and phosphatidylethanolamine plasmalogens in lipid loaded human macrophages. PLoS One. 2018;13 doi: 10.1371/journal.pone.0205706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Numata M, Kandasamy P, Nagashima Y, Fickes R, Murphy RC, Voelker DR. Phosphatidylinositol inhibits respiratory syncytial virus infection. J Lipid Res. 2015;56:578–587. doi: 10.1194/jlr.M055723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petrache I, Natarajan V, Zhen L, et al. Ceramide upregulation causes pulmonary cell apoptosis and emphysema-like disease in mice. Nat Med. 2005;11:491–498. doi: 10.1038/nm1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Courrol LC, de Oliveira Silva FR, Masilamani V. SARS-CoV-2, hemoglobin and protoporphyrin IX: interactions and perspectives. Photodiagn Photodyn Ther. 2021;34 doi: 10.1016/j.pdpdt.2021.102324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su WL, Lin CP, Hang HC, Wu PS, Cheng CF, Chao YC. Desaturation and heme elevation during COVID-19 infection: a potential prognostic factor of heme oxygenase-1. J Microbiol Immunol Infect. 2021;54:113–116. doi: 10.1016/j.jmii.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Firpo MR, Mastrodomenico V, Hawkins GM, et al. Targeting polyamines inhibits coronavirus infection by reducing cellular attachment and entry. ACS Infect Dis. 2021;7:1423–1432. doi: 10.1021/acsinfecdis.0c00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kölker S, Okun JG, Hörster F, et al. 3-Ureidopropionate contributes to the neuropathology of 3-ureidopropionase deficiency and severe propionic aciduria: a hypothesis. J Neurosci Res. 2001;66:666–673. doi: 10.1002/jnr.10012. [DOI] [PubMed] [Google Scholar]
- 33.Mayneris-Perxachs J, Moreno-Navarrete JM, Ballanti M, et al. Lipidomics and metabolomics signatures of SARS-CoV-2 mediators/receptors in peripheral leukocytes, jejunum and colon. Comput Struct Biotechnol J. 2021;19:6080–6089. doi: 10.1016/j.csbj.2021.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abdulrab S, Al-Maweri S, Halboub E. Ursodeoxycholic acid as a candidate therapeutic to alleviate and/or prevent COVID-19-associated cytokine storm. Med Hypotheses. 2020;143 doi: 10.1016/j.mehy.2020.109897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, Maiers JL, Rui Y, Jiang X, Guleng B, Ren J. Apolipoprotein H drives hepatitis B surface antigen retention and endoplasmic reticulum stress during hepatitis B virus infection. Int J Biochem Cell Biol. 2021;131 doi: 10.1016/j.biocel.2020.105906. [DOI] [PubMed] [Google Scholar]
- 36.Su Q, Tsai J, Xu E, et al. Apolipoprotein B100 acts as a molecular link between lipid-induced endoplasmic reticulum stress and hepatic insulin resistance. Hepatology. 2009;50:77–84. doi: 10.1002/hep.22960. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All clinical and multiomics data of deidentified subjects are available in the published appendices; personal health information is protected, as required by law. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD029376 under the project name “Large-scale multiomics analysis identifies novel early markers of COVID-19 severity”; the repository will make the dataset publicly available upon publication and it will be accessible with the dataset identifier. A code snippet that shows the usage of AutoGluon is provided in appendix 1.