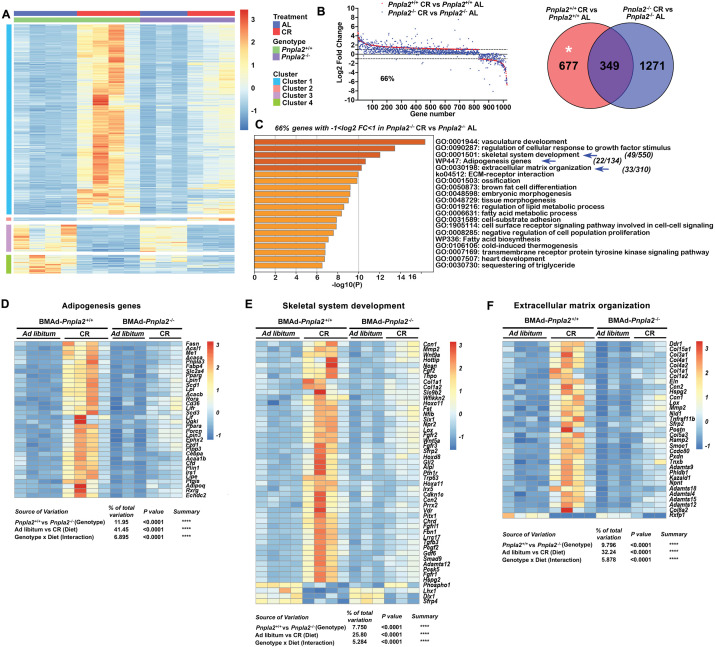
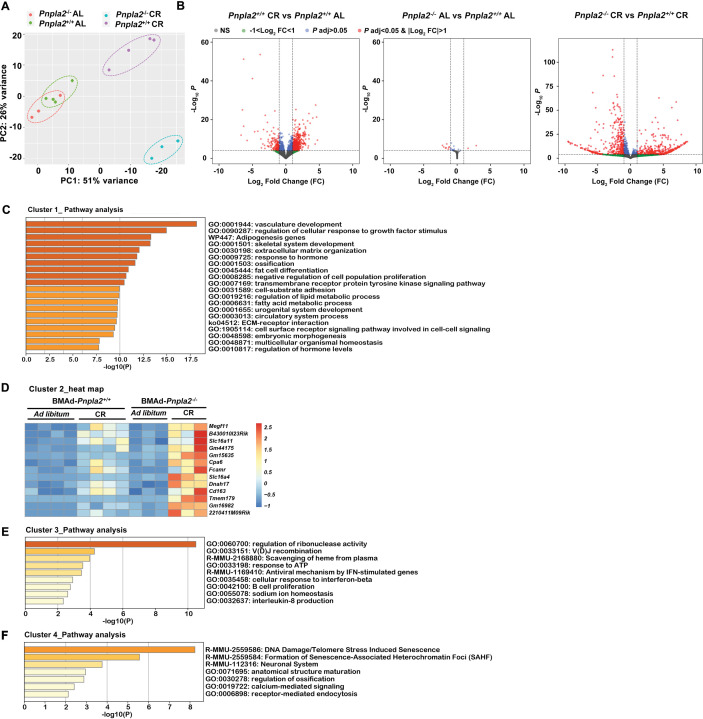
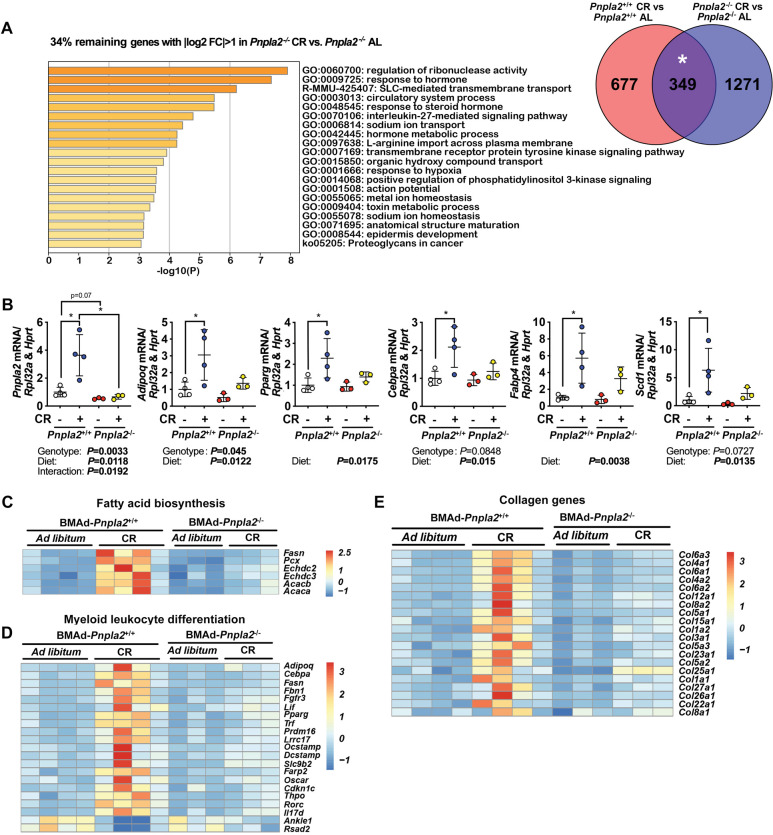
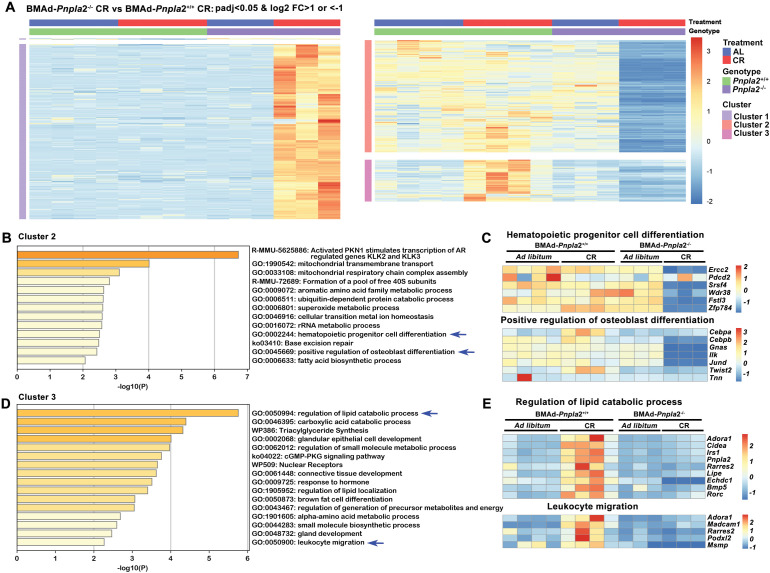
Figure 5. BMAd-Pnpla2 deficiency causes extensive alterations to the bone marrow transcriptome only when coupled with CR.
Male control and BMAd-Pnpla2-/- mice at 24 weeks of age were either fed AL or underwent 30% CR for 6 weeks. Distal tibial cBMAT was flushed and cBMAT from two mice was pooled as one sample for RNAseq analyses (n of 3 or 4 per treatment). (A) Differential genes with our criteria (padjj <0.05 and |Log2 fold change|>1) between BMAd-Pnpla2+/+ CR and BMAd-Pnpla2+/+ad libitum (AL) were grouped into 4 clusters. (B) Genes different between BMAd-Pnpla2+/+ CR and BMAd-Pnpla2+/+ AL were ordered from maximum to minimum log2 fold change (red dots), and compared to corresponding data from BMAd-Pnpla2-/- CR versus BMAd-Pnpla2-/- AL (blue dots). Venn diagram shows the differential genes between BMAd-Pnpla2+/+ CR versus BMAd-Pnpla2+/+ AL BMAT; and BMAd-Pnpla2-/- CR versus BMAd-Pnpla2-/- AL BMAT. (C) Pathway analyses of genes significantly changed by CR in BMAd-Pnpla2+/+ mice, but not in CR mice lacking Pnpla2 (indicated by * area in panel B). Pathways further analyzed by heatmap indicated with blue arrows. (D-F) Expression Z-scores of genes related to adipogenesis (D), skeletal system development (E) and extracellular matrix organization (F) were shown as heatmap. Effects of genotype and diet, and their interactions were analyzed by three-way ANOVA.