Abstract
The glucocorticoid receptor (GR) is a steroid hormone-activated transcription factor that binds to various glucocorticoid response elements to up- or down- regulate the transcription of thousands of genes involved in metabolism, development, stress and inflammatory responses. GR consists of two domains enabling interaction with glucocorticoids, DNA response elements and coregulators, as well as a large intrinsically disordered region that mediates condensate formation. A growing body of structural studies during the past decade have shed new light on GR interactions, providing a new understanding of the mechanisms driving context-specific GR activity. Here, we summarize the established and emerging mechanisms of action of GR, primarily from a structural perspective. This minireview also discusses how the current state of knowledge of GR function may guide future glucocorticoid design with an improved therapeutic index for different inflammatory disorders.
Introduction
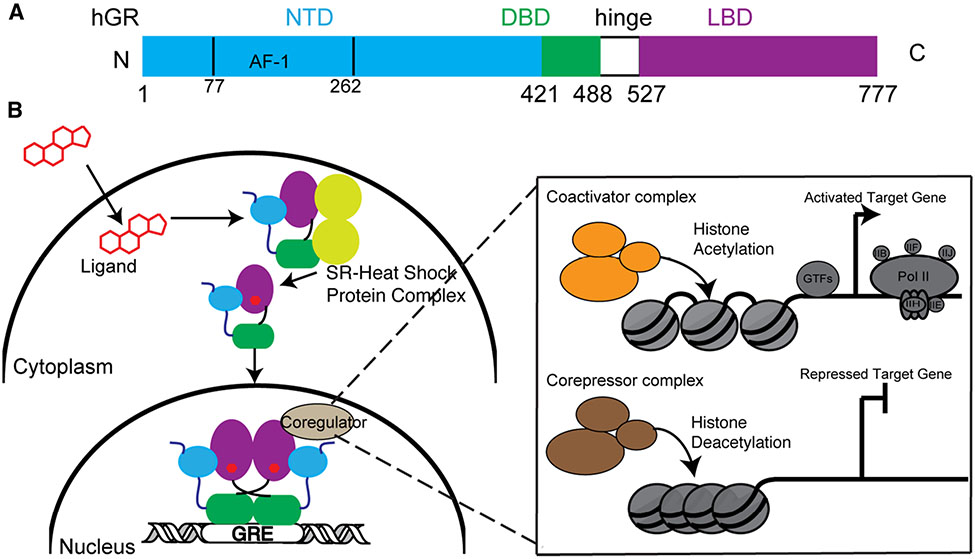
Glucocorticoid receptor (GR) is a member of nuclear receptor (NR) superfamily subgroup 3, which comprises the steroid receptors and includes the mineralocorticoid receptor (MR), progesterone receptor (PR), androgen receptor (AR), and the two estrogen receptors (ERα and ERβ). GR is encoded by the NR subgroup 3 group C member 1 (NR3C1) gene located on chromosome 5 (5q31). It is a ligand-dependent transcription factor sensing the adrenal steroid cortisol to up- or down-regulate thousands of genes influencing metabolism, stress response, development and inflammation [1]. GR is a 777-amino acid multidomain protein containing a largely disordered N-terminal domain (NTD) and two structured domains: a DNA binding domain (DBD), and a C-terminal ligand binding domain (LBD) connected by a flexible hinge region (Figure 1A). The NTD contains an activation function-1 (AF-1) region required for full transcriptional activity. In the absence of ligand, GR is bound to heat shock proteins and sequestered in the cytoplasm. Ligand binding facilitates nuclear translocation where GR binds to specific glucocorticoid response elements (GREs) and recruitment of transcriptional coregulators (co activators and corepressors). The resulting transcriptional complexes are context-specific and lead to either transactivation or transrepression of target genes. Coregulators comprise a diverse group of proteins including histone modifying enzymes such as histone acetyltransferases or deacetylases, but also scaffolding proteins that mediate interactions with other coregulators and the general transcription machinery [2] (Figure 1B). Many seminal structural, biochemical and cellular studies of GR have focused on understanding the molecular details of various aspects of DNA- and ligand-binding. These reductionist strategies have limitations compared with in vivo studies, yet they still shed light on GR’s biological functions and are highlighted below. Recent studies on the roles of NTD in GR activity and condensate formation are also discussed.
Figure 1. Molecular architecture and mechanism of action of glucocorticoid receptor (GR).
(A) Domain architecture of GR highlighting the N-terminal domain (NTD), DNA-binding domain (DBD), hinge region and ligand binding domain (LBD). (B) In the absence of ligand, GR associates with chaperones in the cytosol. Upon ligand binding, GR translocates into nucleus, interacts with its response elements, and recruits coregulators and histone-modifying enzymes to drive gene transactivation and transrepression.
Recognition of various DNA/RNA elements by the GR DBD
DNA binding constrains GR DBD conformational dynamics
The structure of DNA-free GR DBD was first solved in solution by nuclear magnetic resonance (NMR) and revealing two zinc-finger (ZnF) subdomains, each of which coordinates a single zinc ion via four cysteine residues [3, 4]. The subsequent crystal structure of GR DBD bound to a canonical glucocorticoid response element (GRE) showed that the helix following the first ZnF subdomain (ZnF1), referred to as the DNA helix, makes base-specific contacts in the major groove of the hexameric half-sites. ZnF2 contains the dimerization (D)-loop which mediates interactions between the two DBDs on a GRE and is responsible for binding cooperativity. The C-terminus of the core DBD has an additional helix (H3) containing five Lys residues which make additional, nonspecific contacts with the DNA minor groove outside of the recognition sequence [5] (Figure 2A.). Structural comparison between DNA-bound crystal structures and DNA-free NMR structures suggested large conformational changes in the D-loop after DNA binding [6]. A recent DNA-free crystal structure, however, did not show significant changes in the D-loop, but rather identified the lever arm as a hot spot for flexibility [7]. The lever arm, located between the DNA helix and the D-loop, is a flexible loop that had previously been shown to be sensitive to the sequence of bound DNA [5]. The degree of conformational flexibility of the lever arm, including conformational switching between the ‘flipped-in’ and ‘flipped-out’ states of His453, is reduced by DNA binding and DBD dimerization, showing DNA can allosterically alter DBD structure and dynamics.
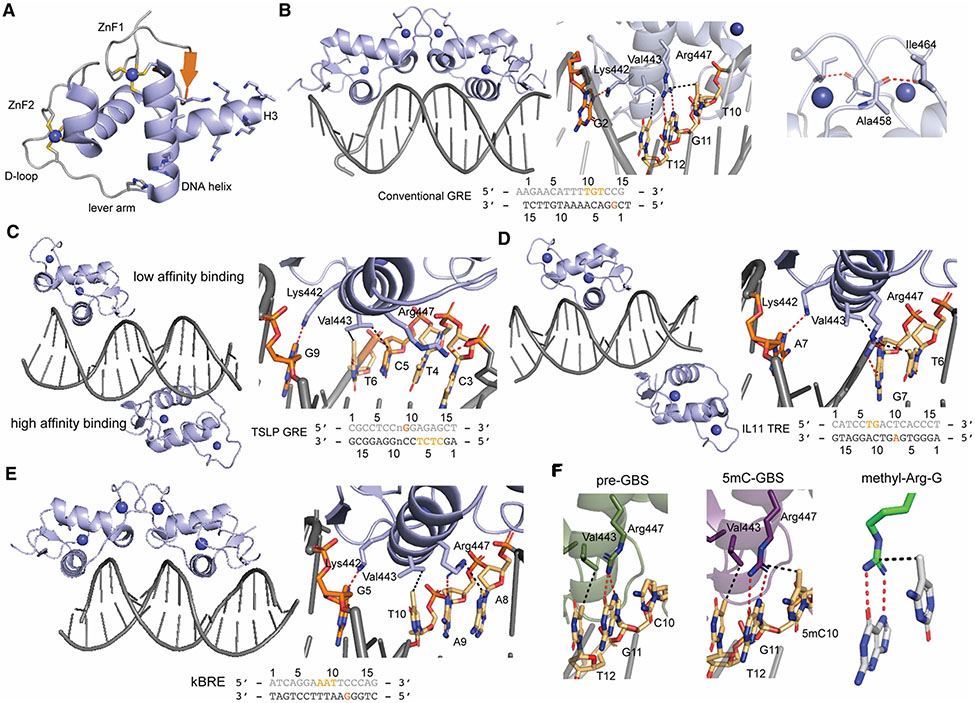
Figure 2. GR DBD interacting with different DNAs.
(A) Overall structure of the apo GR DBD with key subdomains highlighted (PDB: 6CFN). (B) GR DBD bound to conventional GRE as a dimer. Three key residues in the DNA helix form base-specific interaction with nucleotides in both strands (shown in the dark and light orange) from the consensus GRE sequence (middle). Key interactions in the D loop mediate the dimer formation (right) (PDB: 3FYL). (C) GR DBD bound to the TSLP nGRE without a dimer formation (left). Key interactions in the high affinity binding site are highlighted (right) (PDB: 4HN5). (D) GR DBD bound to the IL-11 AP1 response element (left) with only one monomer participating in base-specific interactions (right) (PDB: 5VA7). (E) GR DBD bound to the PLAU NF-kB response element complex(left) with the zoomed base-specific interaction (right) (PDB: 5E6A). (F) GR DBD bound to a pre-GBS (left) and methylated pre-GBS (middle) show the key ‘side-on’ hydrophobic interaction from methylated C (PDB: 6X6D and 6X6E, respectively). The conserved methyl-Arg-G triad is highlighted (right) (PDB: 3C2I). Hydrogen bonds are shown in red while the hydrophobic interactions are shown in black.
Different GRE DNA recognition modes
Canonical GREs are 15 bp in length and consist of two pseudo-palindromic hexameric sites separated by a 3 bp spacer region (5′-A1GAACAnnnTGTTCT15-3′; n, any nucleotide). Three residues in the DNA recognition helix are employed for DNA binding. Arg 447 engages in hydrogen bonds with G11 while Val443 makes hydrophobic contact with T12. Lys442 hydrogen bonds with G2 from the complementary strand. GR Lys442Ala/Arg447Ala is a ‘DNA binding dead’ mutant and frequently used to analyze the role of DNA binding on a particular cellular function [8]. GR binds cooperatively to GRE-containing DNA in a head-to-head fashion, promoting symmetrical interactions within the D-loop (Figure 2B). Mutation of residues within the D-loop weakens cooperative DNA-binding and affects GR activity on a subset of target genes. In particular, the GR Ala458Thr mutant weakens dimerization and has been coined the ‘GRdim’. This Ala458Thr mutant together with Ile634Ala generates what is referred to as the ‘GRmon’, which has been shown to be monomeric in cells [9-11]. Interestingly, recent studies using real-time single molecule number and brightness assays showed that GR forms a dimer of dimers in cells, induced by DNA binding and mediated by both the DBD and LBD [12, 13].
The mechanism behind GR-mediated transactivation is well described, while transrepression of inflammatory genes lacking an apparent GRE is less well understood. In stimulated macrophages for example, GR ChIP-seq peaks contain DNA motifs for activator protein-1 (AP-1) and nuclear factor-kappa-B (NF-κB) with higher abundance than GREs [14]. Interestingly, the same study found binding sites for the interferon regulatory factor 3 (IRF3) associated with repression, but not activation. Originally thought to be solely mediated by DNA-independent protein-protein interaction between GR and AP-1 or NF-κB, later studies have identified a DNA-dependent GR-mediated mechanism for transrepression and evolutionarily conserved negative GREs (nGRE) made up of an inverted repeat with a zero-to-two base spacer [CTCC(n)0-2GGAGA] [15]. Structural studies of GR in complex with the nGRE found upstream of the thymic stromal lymphopoietin (TSLP) promoter showed a head-to-tail binding conformation on opposite sides of DNA, in which the D-loops in the monomers are far away from each other [16]. A two-site (high affinity and low affinity) binding event was observed in the GR-TSLP nGRE interaction contrasting with the cooperative binding of pGRE. In the high affinity binding site, Val443 forms a hydrophobic contact with C5 and T6 while Lys442 forms a hydrogen bond with G9 from the opposite strand. Instead of forming a base-specific hydrogen bond, Arg447 makes hydrophobic contact with T4 and hydrogen bond with base phosphate of C3. There is only one base-specific interaction mediated by Arg447 in the low affinity binding site whereas Lys442 and Val443 do not extend sufficiently deep into the major groove (Figure 2C). Interestingly, other 3-keto steroid receptors, MR, PR and AR that share similar binding to the canonical GRE, are not able to bind an nGRE. Detailed structural and functional analyses show that Gly425Ser substitution along the hGR evolution trajectory alters the backbone conformation of GR and improves its capacity for monomeric nGRE binding [17].
Recent studies found that GR WT but not GR ‘DNA binding Dead’ mutant can occupy AP-1 response elements (TREs) in the absence of AP-1, suggesting GR DNA binding-dependent repression of inflammatory genes beyond the classical model requiring GR-AP-1 interaction [14]. Similar observations were made for the NF-κB response elements (κBREs). Biochemical studies confirmed the direct binding between GR DBD and TRE and κBRE [8, 18]. The structure of GR- interleukin-11 (IL11) TRE complex revealed two GR monomers on the opposite side of TRE, similar to GR-TSLP structure, though one of the monomers does not contribute any base-specific interactions. GR can recognize the consensus TRE sequence of T6GA(G/C)TC11 by DNA helix-mediated interactions: Arg447 makes two hydrogen bonds with G7 and a side-on Van der Waals (VdW) interaction with the methyl group from T6. Val443 forms hydrophobic contact with G7 while Lys442 forms hydrogen bond with A7 from another strand (Figure 2D) [8]. GR DBD/PLAU κBRE structure uncovers the base-specific recognition of A8ATT(T/C)12 consensus sequence, which derived from one DBD monomer although it crystalizes as a dimer, with another monomer only for efficient crystal packing (Figure 2E). Arg447 makes a hydrogen bond with A9 and a hydrophobic interaction with A8. Val443 forms hydrophobic contact with T10 while Lys442 forms hydrogen bond with G5 from another strand [18]. These different nGRE binding modes suggest GR DNA helix can accommodate different DNA sequence via conformational flexibility. Together, these studies show that GR represses a subset of inflammatory genes in a DNA-binding-dependent manner, which is distinct from the DNA-independent tethering mechanism.
Methyl C recognition during GR binding site (GBS) evolution
Cytosine methylation at the 5 position (5mC), occurring commonly at CpG sites, is the most important covalent DNA modification. CpG methylation can produce new transcription factor binding sites during evolution whereby 5mCpG is spontaneously deaminated and fixed as a TpG after DNA replication. Interestingly, the CpG → 5mCpG → TpG transition may have generated a subset of extant GBSs (GnACAnnnTGTnC) [19]. This was shown through a phylogenetic analysis focusing on variation at position 5 within human GBSs (GnACXnnnTGTnC) showing CpG-containing GBSs (GnACGnnnTGTnC) as the most frequent variant in the xenopus, elephant, dog and mouse genomes [19]. These GBSs are coined ‘pre-GBSs’ and may result in functional GBS that are enriched in biological pathways such as muscle function, inflammation and metabolism. Biochemical studies confirmed GR binding to an unmethylated pre-GBS is enhanced upon 5mC methylation. Structural analysis showed CpG methylation generates a favorable VdW contact between Arg447 in the GR DNA helix and the 5-position methyl group of cytosine (mC10), mimicking the interaction observed in the TpG-containing extant GBS (Figure 2A,F). Arg447-mediated hydrogen bonding with a G residue and a ‘side-on’ VdW contact with the preceding methyl-C residue is also observed in GR-TRE recognition (Figure 2D). Indeed, this ‘methyl-Arg-G’ triad is a common mechanism for methylation readout, used by other TFs such as methyl-binding proteins (MBPs), C2H2 zinc finger (ZnF) proteins, p53 and C/EBPβ [19-22] (Figure 2F). Together, this study provides the first genetic and structural evidence of high affinity binding for the likely evolutionary precursor of extant TpG-containing GBS.
RNA recognition
The long non-coding RNA (lncRNA) growth arrest-specific 5 (Gas5) accumulates in growth arrested cells and inhibits cell proliferation via multiple pathways [23-25]. The main Gas5 isoform is 632 nucleotides in length and contains three independent structural modules [26]. The third module (nucleotides 546–566) contains a short stem–loop structure which acts as a GRE mimic (GREM) and suppresses DNA-dependent GR signaling by binding to GR and preventing DNA binding [27]. Currently there is no available structure of the GR/Gas5 GREM complex. Hydroxyl radical footprinting coupled with NMR titration experiments identified the Gas5 nucleotides contacting GR and confirmed that Gas5 engages GR at its DNA binding residues. The D-loop does not participate in Gas5 binding suggesting no DBD dimer formation on RNA [28]. GR uses its DNA helix to penetrate a widened major groove formed in GREM, with the crucial interaction as the Arg447 and G549. G549A mutation caused steric clashes with Arg447, abolished the GR binding, and prevented Gas5-induced apoptosis [28]. Interestingly, a recent study found contribution of H3 of GR DBD in Gas5 interaction based on solution NMR and Ala scanning studies [29]. GR can bind to a diverse range of hairpin motifs suggesting that other RNAs, e.g lncRNAs or nascent RNAs may be able to regulate GR function [29].
GR LBD structures
Overall LBD structure
The first GR LBD structure was determined with the potent agonist dexamethasone (Figure 3A) and a peptide derived from co-activator Tif2 [30]. Overall, GR LBD adopts the classic NR/SR structure and forms an α helical sandwich with three layers, consisting of 11 α helices and 4 small β strands. Helix 1(H1) and H3 form the front layer of the helical sandwich; H7 and H10 form the back layer, whereas H4, H5, H8 and H9 form the middle layer (Figure 3B). At the base of the LBD helices, sits the ligand binding cavity which specifically recognizes glucocorticoids through extensive hydrophobic interactions and a series of hydrogen bonds with the carbonyl or hydroxyl groups attached to the steroid scaffold (Figure 3A). The C3-carbonyl forms a water mediated hydrogen bond network with Gln570 and Arg611, which is critical to maintain the ligand in the correct orientation [31]. The C17-hydroxyl on the D ring hydrogen bonds with Gln642 whereas the hydroxyls O4 and O5 hydrogen bond with Thr739. Residue Asn564 hydrogen bonds with both C11- and C21-hydroxyls (Figure 3C). These interactions are subject to changes with modifications of the steroidal core (see below).
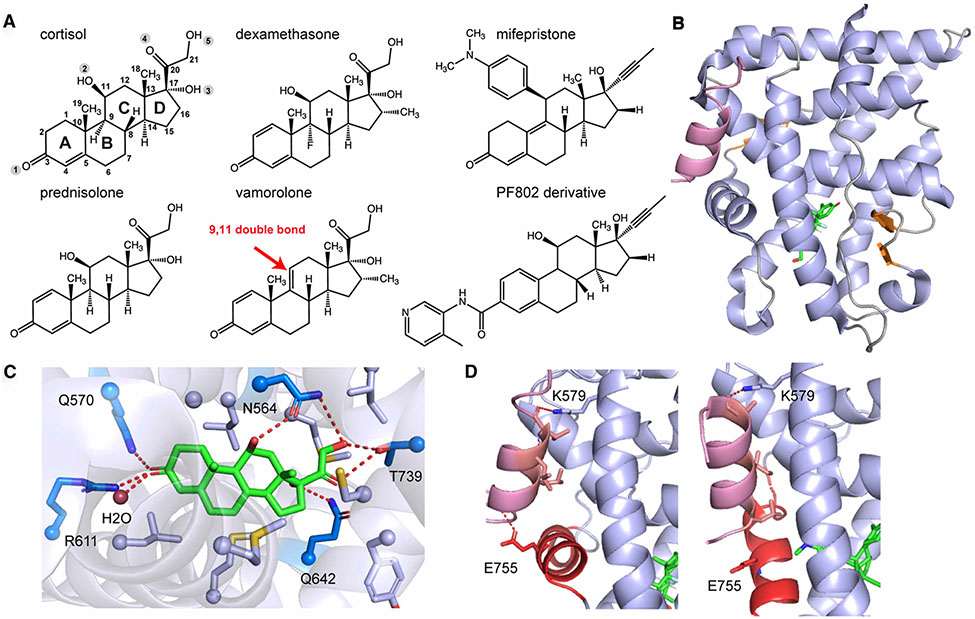
Figure 3. GR LBD interacting with ligands and coregulators.
(A). Chemical structures of common GR ligands with the canonical numbering shown on the structure of cortisol. (B). Overall structure of GR LBD with cortisol (green) bound to Tif2 (light purple), with α-helices shown in light blue, β-strands in orange, and loops in gray (PDB: 1M2Z). (C) Extensive hydrogen bonds (dark blue residues) and hydrophobic interactions (light blue residues) are formed to accommodate cortisol binding. (D) Different GR coregulator binding modes. In the presence of cortisol (agonist), the LxxLL motif from the co-activator forms hydrophobic contacts with GR residues in the AF-2 site. Primary charge clamps mediated by Lys579 (from H3) and Glu755 (from AF-H) hold the peptide in place. In the presence of mifepristone (antagonist), the AF-H is displaced from the agonist-bound position, disrupting interaction with the charge clamp residues Glu755 (PDB: 3H52).
The TIF2-derived co-activator peptide contains a conserved LxxLL (x, any amino acid) motif that forms a short alpha helix. These leucines are oriented on one face of this helix and make hydrophobic interactions with a groove located on the AF-2 surface formed by H3, H4, and H12 (also known as the activation function helix, AF-H). GR Glu755 from AF-H and Lys579 from H3 form the hydrogen bonds with the N- and C- termini, respectively, of the co-activator peptide creating charge-clamps that further strengthens co-activator binding (Figure 3D). Sequence variations present in coregulators flanking the LxxLL motif can influence binding and are thought to regulate unique downstream signaling, as shown in the structural comparison between GR-dexamethasone–Tif2 and –PGC1a complexes [32]. Disruption of the AF-2 through an engineered steric clash is accomplished by the antagonist mifepristone (also known as RU-486) has been used for the treatment of Cushing’s syndrome, depression and psychosis, neuropathic pain and glaucoma [33-35] (Figure 3A). Structural studies of GR in complex with mifepristone showed high conformational flexibility of the AF-H and the pre-AF-H loop observing that the AF-H can stack into the co-activator binding groove to antagonize gene transactivation [36]. Another structural study using the nuclear receptor corepressor (NCoR) peptide, known to interact with GR in the presence of mifepristone, showed hydrophobic interactions with the conserved Lxxx(I/L)xxx(I/L) motif found in corepressors and the charge clamp formed with Lys579. Displacement of the AF-H from the agonist-bound conformation, caused by the C11 modification, disabled the formation of Glu755-mediated charge-clamp [37] (Figure 3D).
Recent development of selective GR modulators
The endogenous glucocorticoid cortisol (also known as hydrocortisone) has been clinically used as an anti-inflammatory agent by repressing gene transcription, i.e. transrepression [38] (Figure 3A). Synthetic glucocorticoids, such as dexamethasone, mometasone furoate, budesonide and triamcinolone acetonide, have better therapeutic effects than hydrocortisone for inflammatory and autoimmune disease, but similar to hydrocortisone, they can produce adverse side effects such as weight gain, Cushing’s syndrome, and osteoporosis, attributed to GR-mediated gene transactivation [39]. Selective glucocorticoid receptor modulators (SGRMs, previously known as dissociated glucocorticoids) that preserve the anti-inflammatory effects but without the debilitating side effects hold promise for future of glucocorticoids-based treatment [40]. Different glucocorticoids have been developed by different modifications on the steroidal core. PF806 is an SGRM developed by Pfizer currently in clinical trials for rheumatoid arthritis [41, 42]. A recent study synthesized a series of PF806 derivatives with modifications at C11 and C17 to drive antagonism and improve the drug potency, respectively, sharing similar molecular mechanisms of existing drugs mifepristone and mometasone furoate [43] (Figure 3A). Additional substitutions at C3 extend to the C terminus of H5, open up the ligand binding pocket and can allosterically alter the conformation of N terminus of H3 and pre-AF-H loop as previously shown by the GR LBD structure with deacylcortivazol [44]. Molecular dynamics simulations coupled with network analysis found different communication pathways between H5 and AF-H in C3-modified glucocorticoids from dexamethasone, correlated with the distance between H3 and H11. This structure-activity effort reveals the GR signaling mechanism and connection between different GR-controlled physiological effects including anti-inflammation effects, glucose disposal and muscle atrophy [43]. Other SGRMs from structure-based design achieved highly potent glucocorticoids by combining C3, C17 and C21 substitutions with better effects on repressing lung and neutrophilic airway inflammation than dexamethasone and fluticasone furoate but reduced side effects, which can be used for the future asthma treatment [45-47]. In parallel with this, nonsteroidal GR modulators have also been designed to dissociate GR transactivation from transrepression for better treatment of pulmonary disease and joint inflammation [48, 49].
Δ-9,11 steroids with much improved side effect profiles
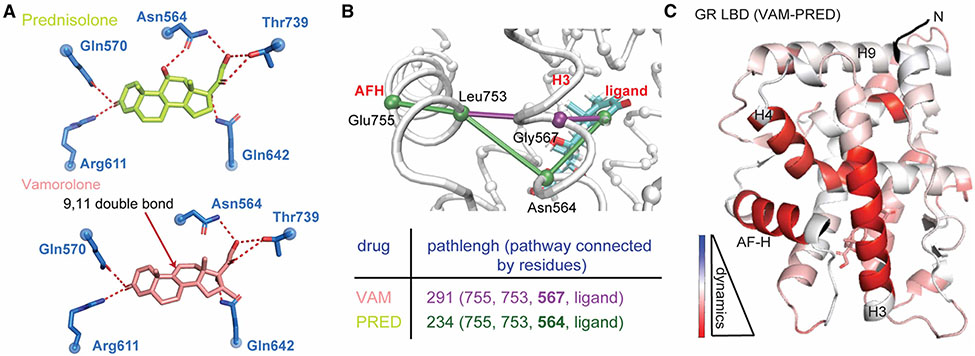
Rather than adding bulky adducts to the steroidal core as mentioned above, subtle modification of the steroid backbone can have great effects towards better SGRMs. Vamorolone (previously known as VBP15) is the lead Δ-9,11 steroid that has superior side effect profile compared with conventional glucocorticoids [50]. It maintains the anti-inflammatory effects on colitis, allergic lung inflammation and asthma and is currently in phase 2b clinical trial for Duchenne muscular dystrophy (DMD) [51-54]. Vamorolone is structurally similar to other glucocorticoids but has an additional double bond between C9 and C11 and thus lacks the C11 hydroxyl group. It has high affinity for GR binding but does not induce classical GR transactivation as opposed to prednisolone, the current FDA-approved DMD treatment [55]. Structures of GR-ligand complexes showed that the hydrogen bond formed between Asn564 and the C11-hydroxyl in prednisolone is not present in GR’s interaction with vamorolone (Figure 4A). Asn564 was identified as the crucial residue in H3 connecting ligand binding to the AF-2 region (Figure 4B). Vamorolone induced weaker allosteric communication to the AF-2 region and more pronounced conformational dynamics compared with prednisolone (Figure 4C). Disruption of this allosteric network by removal of a single hydrogen bond selectively weakens the binding with co-activators but not corepressors and thus reduces GR-driven transactivation while leaving transrepression intact [56]. These clinical and mechanistic studies suggest incorporation of a 9,11-double bond into current steroidal core could be remarkably beneficial in driving dissociation of GR effects and improve the therapeutic index of future drugs.
Figure 4. Vamorolone reduces unwanted GR activity by disrupting a key hydrogen bond network that supports transactivation.
(A) A critical hydrogen bond formed between GR Asn564 and C11-OH of prednisolone (top) is lacking when vamorolone (bottom) binds. (B) Ligand-H12 allostery is rewired and weakened by Vamolone relative to a glucocorticoid bypassing the critical and conserved residue Asn564. (C) HDX shows that the GR conformation is more dynamic in solution when in complex with Vamorolone relative to the strong agonist prednisolone. The AF-2 site in particular is highly destabilized due to an inability of Vamorolone to support strong communication from the ligand binding pocket to this co-regulator interaction surface.
Intrinsic disorder in the NTD
The NTD is 420-residues in length and harbors most of GR’s post-translational modification (PTM) sites that have been identified so far, such as two clustered phosphorylation sites from Ser113 to Ser141 and from Ser203 to Ser226 [57]. These phosphorylations can affect transcriptional outcome in a gene-specific and cell-type-specific manner [58, 59]. The NTD is largely disordered, which has impeded structural characterization. Thermodynamics analyses suggest the presence of two regions, a functional region (residue 98–420) that contains the AF-1 and a regulatory region (residue 1-97) [60]. GR translational isoforms generated from alternative start codons and thus containing distinct truncated versions of the NTD exhibit very different transcriptional activities, indicating the regulatory region can tune GR activity allosterically. These resultant isoforms utilize different intrinsically disordered regulatory regions and work in concert with the different PTMs to fine-tune GR activity under physiological contexts [61].
An emerging role of intrinsically disordered regions (IDRs) is the formation of biological condensates via weak, multivalent interactions. While there is no consensus yet in the broader scientific community about the functional role of biological condensates, tantalizing new data suggest that transcriptional condensates containing transcription factors, co-regulators, and the transcription machinery are associated with gene activation [62, 63]. GR has been shown to form phase-separated condensates that contain the known GR co-activators MED1 and NCoA2 in the nucleus [64]. More recently, single molecule tracking experiments combined with a machine learning classification approach found a population of GR molecules in a confined state with reduced mobility that is not chromatin associated [65]. Consistent with the role of IDRs in condensate formation, the presence of this confined state depends on the GR NTD. Interestingly, motif analysis from ChIP-Seq experiments showed that GR lacking the NTD has a higher preference for binding to GREs compared with the full-length protein, suggesting that localization to condensates via the N-terminal IDR facilitates GR binding to a broader collection of sequences including lower affinity binding sites.
A study using recombinant full-length GR showed that it can form condensates in vitro which specifically enrich transcriptional co-regulator proteins via their IDRs [66]. LxxLL motifs are utilized for recruitment into condensates to different extents by different coregulators suggesting a differential ability to sense and respond to GR drug binding. Importantly, binding to activating, but not repressive DNAs resulted in enhanced recruitment of co-activators and reduced recruitment of co-repressors to GR condensates.
Together these recent studies suggest that condensation is an important process for modulating GR function and may present novel avenues for therapeutic intervention. More work needs to be done to define the role of condensation in DNA-dependent and -independent transcriptional activities, in coregulator recruitment, and ultimately the context-dependent behavior of GR. Does phase separation and the selective recruitment of coregulators in different DNA-binding states contribute to cell type- and gene-specific activities of GR?
Perspectives.
The importance of the field: Cortisol and other widely prescribed glucocorticoids remain the most efficacious treatment for inflammatory diseases by acting on GR to achieve their therapeutic effects. However, their on-target interaction with GR leads to dose-limiting side effects due to a rich and complicated array of GR functions, particularly context-dependent gene transcription.
A summary of the current thinking: Accumulated knowledge of GR structure has been pivotal in establishing our current view of the comprehensive GR signaling and how it drives gene transactivation/ transrepression by binding to different GREs and coregulators in response to different ligands. Recent understanding of the intrinsically disordered NTD add an additional layer of complexity by revealing the role of condensation in modulating GR function.
A comment on future directions: While most studies so far only provided structural information of GR individual domains, the forthcoming structure of full-length GR in complex with GREs and coregulators, particularly with the recent advances in cryo-electron microscope technique and its successful application in determining the structures of other full-length nuclear receptor complexes [67-69], will shed tremendous light on how each GR domain fits together to achieve its remarkable biological function.
Funding
X.L. was supported by research grant from Foundation to Eradicate Duchenne. E.A.O was supported by grant R01DK115213 from the NIH National Institute of Diabetes and Digestive and Kidney Diseases.
Abbreviations
- AF
activation function
- DBD
DNA binding domain
- GBS
GR binding site
- GR
glucocorticoid receptor
- GRE
glucocorticoid response element
- IDR
intrinsically disordered regions
- LBD
ligand binding domain
- NR
nuclear receptor
- NTD
N-terminal domain
- SGRM
selective glucocorticoid receptor modulators
- ZnF
zinc-finger
Footnotes
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Kadmiel M and Cidlowski JA (2013) Glucocorticoid receptor signaling in health and disease. Trends Pharmacol. Sci 34, 518–530 10.1016/j.tips.2013.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartlett AA, Lapp HE and Hunter RG (2019) Epigenetic mechanisms of the glucocorticoid receptor. Trends Endocrinol. Metab 30, 807–818 10.1016/jtem.2019.07.003 [DOI] [PubMed] [Google Scholar]
- 3.Hard T, Kellenbach E, Boelens R, Maler BA, Dahlman K, Freedman LP et al. (1990) Solution structure of the glucocorticoid receptor DNA-binding domain. Science 249, 157–160 10.1126/science.2115209 [DOI] [PubMed] [Google Scholar]
- 4.Baumann H, Paulsen K, Kovacs H, Berglund H, Wright AP, Gustafsson JA et al. (1993) Refined solution structure of the glucocorticoid receptor DNA-binding domain. Biochemistry 32, 13463–13471 10.1021/bi00212a011 [DOI] [PubMed] [Google Scholar]
- 5.Meijsing SH, Pufall MA, So AY, Bates DL, Chen L and Yamamoto KR (2009) DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 324, 407–410 10.1126/science.1164265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luisi BF, Xu WX, Otwinowski Z, Freedman LP, Yamamoto KR and Sigler PB (1991) Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature 352, 497–505 10.1038/352497a0 [DOI] [PubMed] [Google Scholar]
- 7.Frank F, Okafor CD and Ortlund EA (2018) The first crystal structure of a DNA-free nuclear receptor DNA binding domain sheds light on DNA-driven allostery in the glucocorticoid receptor. Sci. Rep 8, 13497 10.1038/s41598-018-31812-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weikum ER, de Vera IMS, Nwachukwu JC, Hudson WH, Nettles KW, Kojetin DJ et al. (2017) Tethering not required: the glucocorticoid receptor binds directly to activator protein-1 recognition motifs to repress inflammatory genes. Nucleic Acids Res. 45, 8596–8608 10.1093/nar/gkx509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jewell CM, Scoltock AB, Hamel BL, Yudt MR and Cidlowski JA (2012) Complex human glucocorticoid receptor dim mutations define glucocorticoid induced apoptotic resistance in bone cells. Mol. Endocrinol 26, 244–256 10.1210/me.2011-1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Presman DM, Ogara MF, Stortz M, Alvarez LD, Pooley JR, Schiltz RL et al. (2014) Live cell imaging unveils multiple domain requirements for in vivo dimerization of the glucocorticoid receptor. PLoS Biol. 12, e1001813 10.1371/journal.pbio.1001813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Louw A (2019) GR dimerization and the impact of GR dimerization on GR protein stability and half-life. Front. Immunol 10, 1693 10.3389/fimmu.2019.01693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paakinaho V, Johnson TA, Presman DM and Hager GL (2019) Glucocorticoid receptor quaternary structure drives chromatin occupancy and transcriptional outcome. Genome Res. 29, 1223–1234 10.1101/gr.244814.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Presman DM, Ganguly S, Schiltz RL, Johnson TA, Karpova TS and Hager GL (2016) DNA binding triggers tetramerization of the glucocorticoid receptor in live cells. Proc. Natl Acad. Sci. U.S.A 113, 8236–8241 10.1073/pnas.1606774113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uhlenhaut NH, Barish GD, Yu RT, Downes M, Karunasiri M, Liddle C et al. (2013) Insights into negative regulation by the glucocorticoid receptor from genome-wide profiling of inflammatory cistromes. Mol. Cell 49, 158–171 10.1016/j.molcel.2012.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Surjit M, Ganti KP, Mukherji A, Ye T, Hua G, Metzger D et al. (2011) Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell 145, 224–241 10.1016/j.cell.2011.03.027 [DOI] [PubMed] [Google Scholar]
- 16.Hudson WH, Youn C and Ortlund EA (2013) The structural basis of direct glucocorticoid-mediated transrepression. Nat. Struct. Mol. Biol 20, 53–58 10.1038/nsmb.2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hudson WH, Kossmann BR, de Vera IM, Chuo SW, Weikum ER, Eick GN et al. (2016) Distal substitutions drive divergent DNA specificity among paralogous transcription factors through subdivision of conformational space. Proc. Natl Acad. Sci. U.S.A 113, 326–331 10.1073/pnas.1518960113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hudson WH, Vera IMS, Nwachukwu JC, Weikum ER, Herbst AG, Yang Q et al. (2018) Cryptic glucocorticoid receptor-binding sites pervade genomic NF-kappaB response elements. Nat. Commun 9, 1337 10.1038/s41467-018-03780-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu X, Weikum ER, Tilo D, Vinson C and Ortlund EA (2021) Structural basis for glucocorticoid receptor recognition of both unmodified and methylated binding sites, precursors of a modern recognition element. Nucleic Acids Res. 49, 8923–8933 10.1093/nar/gkab605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hodges AJ, Hudson NO and Buck-Koehntop BA (2020) Cys2his2 zinc finger methyl-CpG binding proteins: getting a handle on methylated DNA. J. Mol. Biol 432, 1640–1660 10.1016/jjmb.2019.09.012 [DOI] [PubMed] [Google Scholar]
- 21.Kribelbauer JF, Lu XJ, Rohs R, Mann RS and Bussemaker HJ (2019) Toward a mechanistic understanding of DNA methylation readout by transcription factors. J. Mol. Biol, 10.1016/jjmb.2019.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang J, Horton JR, Wang D, Ren R, Li J, Sun D et al. (2019) Structural basis for effects of CpA modifications on C/EBPbeta binding of DNA. Nucleic Acids Res. 47, 1774–1785 10.1093/nar/gky1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mourtada-Maarabouni M, Hedge VL, Kirkham L, Farzaneh F and Williams GT (2008) Growth arrest in human T-cells is controlled by the non-coding RNA growth-arrest-specific transcript 5 (GAS5). J. Cell Sci 121, 939–946 10.1242/jcs.024646 [DOI] [PubMed] [Google Scholar]
- 24.Mourtada-Maarabouni M, Hasan AM, Farzaneh F and Williams GT (2010) Inhibition of human T-cell proliferation by mammalian target of rapamycin (mTOR) antagonists requires noncoding RNA growth-arrest-specific transcript 5 (GAS5). Mol. Pharmacol 78, 19–28 10.1124/mol.110.064055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pickard MR, Mourtada-Maarabouni M and Williams GT (2013) Long non-coding RNA GAS5 regulates apoptosis in prostate cancer cell lines. Biochim. Biophys. Acta 1832, 1613–1623 10.1016/j.bbadis.2013.05.005 [DOI] [PubMed] [Google Scholar]
- 26.Frank F, Kavousi N, Bountali A, Dammer EB, Mourtada-Maarabouni M and Ortlund EA (2020) The lncRNA growth arrest specific 5 regulates cell survival via distinct structural modules with independent functions. Cell Rep. 32, 107933 10.1016/j.celrep.2020.107933 [DOI] [PubMed] [Google Scholar]
- 27.Kino T, Hurt DE, Ichijo T, Nader N and Chrousos GP (2010) Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal 3, ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hudson WH, Pickard MR, de Vera IM, Kuiper EG Mourtada-Maarabouni M Conn GL et al. (2014) Conserved sequence-specific lincRNA-steroid receptor interactions drive transcriptional repression and direct cell fate. Nat. Commun 5, 5395 10.1038/ncomms6395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parsonnet NV, Lammer NC, Holmes ZE, Batey RT and Wuttke DS (2019) The glucocorticoid receptor DNA-binding domain recognizes RNA hairpin structures with high affinity. Nucleic Acids Res. 47, 8180–8192 10.1093/nar/gkz486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bledsoe RK, Montana VG, Stanley TB, Delves CJ, Apolito CJ, McKee DD et al. (2002) Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell 110, 93–105 10.1016/S0092-8674(02)00817-6 [DOI] [PubMed] [Google Scholar]
- 31.Weikum ER, Okafor CD, D’Agostino EH, Colucci JK and Ortlund EA (2017) Structural analysis of the glucocorticoid receptor ligand-binding domain in complex with triamcinolone acetonide and a fragment of the atypical coregulator, small heterodimer partner. Mol. Pharmacol 92, 12–21 10.1124/mol.117.108506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X, Wang Y and Ortlund EA (2019) First high-resolution crystal structures of the glucocorticoid receptor ligand-binding domain-peroxisome proliferator-activated gamma coactivator 1-alpha complex with endogenous and synthetic glucocorticoids. Mol. Pharmacol 96, 408–417 10.1124/mol.119.116806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fleseriu M, Biller BM, Findling JW, Molitch ME, Schteingart DE, Gross C et al. (2012) Mifepristone, a glucocorticoid receptor antagonist, produces clinical and metabolic benefits in patients with cushing’s syndrome. J. Clin. Endocrinol. Metab 97, 2039–2049 10.1210/jc.2011-3350 [DOI] [PubMed] [Google Scholar]
- 34.Gallagher P and Young AH (2006) Mifepristone (RU-486) treatment for depression and psychosis: a review of the therapeutic implications. Neuropsychiatr. Dis. Treat 2, 33–42 [PMC free article] [PubMed] [Google Scholar]
- 35.Clark RD (2008) Glucocorticoid receptor antagonists. Curr. Top. Med. Chem 8, 813–838 10.2174/156802608784535011 [DOI] [PubMed] [Google Scholar]
- 36.Kauppi B, Jakob C, Farnegardh M, Yang J, Ahola H, Alarcon M et al. (2003) The three-dimensional structures of antagonistic and agonistic forms of the glucocorticoid receptor ligand-binding domain: RU-486 induces a transconformation that leads to active antagonism. J. Biol. Chem 278, 22748–22754 10.1074/jbc.M212711200 [DOI] [PubMed] [Google Scholar]
- 37.Schoch GA, D’Arcy B, Stihle M, Burger D, Bar D, Benz J et al. (2010) Molecular switch in the glucocorticoid receptor: active and passive antagonist conformations. J. Mol. Biol 395, 568–577 10.1016/jjmb.2009.11.011 [DOI] [PubMed] [Google Scholar]
- 38.Clark AR and Belvisi MG (2012) Maps and legends: the quest for dissociated ligands of the glucocorticoid receptor. Pharmacol. Ther 134, 54–67 10.1016/j.pharmthera.2011.12.004 [DOI] [PubMed] [Google Scholar]
- 39.Schacke H, Docke WD and Asadullah K (2002) Mechanisms involved in the side effects of glucocorticoids. Pharmacol. Ther 96, 23–43 10.1016/S0163-7258(02)00297-8 [DOI] [PubMed] [Google Scholar]
- 40.Schacke H, Rehwinkel H and Asadullah K (2005) Dissociated glucocorticoid receptor ligands: compounds with an improved therapeutic index. Curr. Opin. Investig. Drugs 6, 503–507 [PubMed] [Google Scholar]
- 41.Hu X, Du S, Tunca C, Braden T, Long KR, Lee J et al. (2011) The antagonists but not partial agonists of glucocorticoid receptor ligands show substantial side effect dissociation. Endocrinology 152, 3123–3134 10.1210/en.2010-1447 [DOI] [PubMed] [Google Scholar]
- 42.Stock T, Fleishaker D, Wang X, Mukherjee A and Mebus C (2017) Improved disease activity with fosdagrocorat (PF-04171327), a partial agonist of the glucocorticoid receptor, in patients with rheumatoid arthritis: a phase 2 randomized study. Int. J. Rheum. Dis 20, 960–970 10.1111/1756-185X.13053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bruno NE, Nwachukwu JC, Srinivasan S, Nettles CC, Izard T, Jin Z et al. (2021) Chemical systems biology reveals mechanisms of glucocorticoid receptor signaling. Nat. Chem. Biol 17, 307–316 10.1038/s41589-020-00719-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suino-Powell K, Xu Y, Zhang C, Tao YG, Tolbert WD, Simons SS Jr et al. (2008) Doubling the size of the glucocorticoid receptor ligand binding pocket by deacylcortivazol. Mol. Cell. Biol 28, 1915–1923 10.1128/MCB.01541-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He Y, Shi J, Nguyen QT, You E, Liu H, Ren X et al. (2019) Development of highly potent glucocorticoids for steroid-resistant severe asthma. Proc. Natl Acad. Sci. U.S.A 116, 6932–6937 10.1073/pnas.1816734116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He Y, Shi J, Yi W, Ren X, Gao X, Li J et al. (2015) Discovery of a highly potent glucocorticoid for asthma treatment. Cell Discov. 1, 15035 10.1038/celldisc.2015.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He Y, Yi W, Suino-Powell K, Zhou XE, Tolbert WD, Tang X et al. (2014) Structures and mechanism for the design of highly potent glucocorticoids. Cell Res. 24, 713–726 10.1038/cr.2014.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ripa L, Edman K, Dearman M, Edenro G, Hendrickx R, Ullah V et al. (2018) Discovery of a novel oral glucocorticoid receptor modulator (AZD9567) with improved side effect profile. J. Med. Chem 61, 1785–1799 10.1021/acs.jmedchem.7b01690 [DOI] [PubMed] [Google Scholar]
- 49.Hemmerling M, Nilsson S, Edman K, Eirefelt S, Russell W, Hendrickx R et al. (2017) Selective nonsteroidal glucocorticoid receptor modulators for the inhaled treatment of pulmonary diseases. J. Med. Chem 60, 8591–8605 10.1021/acs.jmedchem.7b01215 [DOI] [PubMed] [Google Scholar]
- 50.Reeves EKM, Hoffman EP, Nagaraju K, Damsker JM and McCall JM (2013) VBP15: preclinical characterization of a novel anti-inflammatory delta 9,11 steroid. Bioorg. Med. Chem 21, 2241–2249 10.1016/j.bmc.2013.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Damsker JM, Conklin LS, Sadri S, Dillingham BC, Panchapakesan K, Heier CR et al. (2016) VBP15, a novel dissociative steroid compound, reduces NFkappaB-induced expression of inflammatory cytokines in vitro and symptoms of murine trinitrobenzene sulfonic acid-induced colitis. Inflamm. Res 65, 737–743 10.1007/s00011-016-0956-8 [DOI] [PubMed] [Google Scholar]
- 52.Damsker JM, Dillingham BC, Rose MC, Balsley MA, Heier CR, Watson AM et al. (2013) VBP15, a glucocorticoid analogue, is effective at reducing allergic lung inflammation in mice. PLoS ONE 8, e63871 10.1371/journal.pone.0063871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Conklin LS, Damsker JM, Hoffman EP, Jusko WJ, Mavroudis PD, Schwartz BD et al. (2018) Phase IIa trial in duchenne muscular dystrophy shows vamorolone is a first-in-class dissociative steroidal anti-inflammatory drug. Pharmacol. Res 136, 140–150 10.1016/j.phrs.2018.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Freishtat RJ, Nino G, Tsegaye Y, Alcala SE, Benton AS, Watson AM et al. (2015) Pharmacologically-induced mitotic synchrony in airway epithelial cells as a mechanism of action of anti-inflammatory drugs. Respir. Res 16, 132 10.1186/s12931-015-0293-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heier CR, Damsker JM, Yu Q, Dillingham BC, Huynh T, Van der Meulen JH et al. (2013) VBP15, a novel anti-inflammatory and membrane-stabilizer, improves muscular dystrophy without side effects. EMBO Mol. Med 5, 1569–1585 10.1002/emmm.201302621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu X, Wang Y, Gutierrez JS, Damsker JM, Nagaraju K, Hoffman EP et al. (2020) Disruption of a key ligand-H-bond network drives dissociative properties in vamorolone for duchenne muscular dystrophy treatment. Proc. Natl Acad. Sci. U.S.A 117, 24285–24293 10.1073/pnas.2006890117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oakley RH and Cidlowski JA (2011) Cellular processing of the glucocorticoid receptor gene and protein: new mechanisms for generating tissue-specific actions of glucocorticoids. J. Biol. Chem 286, 3177–3184 10.1074/jbc.R110.179325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hay CW, Shanley L, Davidson S, Cowie P, Lear M, McGuffln P et al. (2014) Functional effects of polymorphisms on glucocorticoid receptor modulation of human anxiogenic substance-P gene promoter activity in primary amygdala neurones. Psychoneuroendocrinology 47, 43–55 10.1016/j.psyneuen.2014.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weikum ER, Knuesel MT, Ortlund EA and Yamamoto KR (2017) Glucocorticoid receptor control of transcription: precision and plasticity via allostery. Nat. Rev. Mol. Cell Biol 18, 159–174 10.1038/nrm.2016.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li J, Motlagh HN, Chakuroff C, Thompson EB and Hilser VJ (2012) Thermodynamic dissection of the intrinsically disordered N-terminal domain of human glucocorticoid receptor. J. Biol. Chem 287, 26777–26787 10.1074/jbc.M112.355651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li J, White JT, Saavedra H, Wrabl JO, Motlagh HN, Liu K et al. (2017) Genetically tunable frustration controls allostery in an intrinsically disordered transcription factor. eLife 6, e30688 10.7554/eLife.30688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K et al. (2018) Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 10.1126/science.aar3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boija A, Klein IA, Sabari BR, Dall’Agnese A, Coffey EL, Zamudio AV et al. (2018) Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842–55 e16 10.1016/j.cell.2018.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stortz M, Pecci A, Presman DM and Levi V (2020) Unraveling the molecular interactions involved in phase separation of glucocorticoid receptor. BMC Biol. 18, 59 10.1186/s12915-020-00788-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Garcia DA, Johnson TA, Presman DM, Fettweis G, Wagh K, Rinaldi L et al. (2021) An intrinsically disordered region-mediated confinement state contributes to the dynamics and function of transcription factors. Mol. Cell 81, 1484–98 e6 10.1016/j.molcel.2021.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Frank F, Liu X and Ortlund EA (2021) Glucocorticoid receptor condensates link DNA-dependent receptor dimerization and transcriptional transactivation. Proc. Natl Acad. Sci. U.S.A 118, e2024685118 10.1073/pnas.2024685118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu X, Yi P, Hamilton RA, Shen H, Chen M, Foulds CE et al. (2020) Structural insights of transcriptionally active, full-length androgen receptor coactivator complexes. Mol. Cell 79, 812–23 e4 10.1016/j.molcel.2020.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yi P, Wang Z, Feng Q, Pintilie GD, Foulds CE, Lanz RB et al. (2015) Structure of a biologically active estrogen receptor-coactivator complex on DNA. Mol. Cell 57, 1047–1058 10.1016/j.molcel.2015.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Orlov I, Rochel N, Moras D and Klaholz BP (2012) Structure of the full human RXR/VDR nuclear receptor heterodimer complex with its DR3 target DNA. EMBOJ. 31, 291–300 10.1038/emboj.2011.445 [DOI] [PMC free article] [PubMed] [Google Scholar]