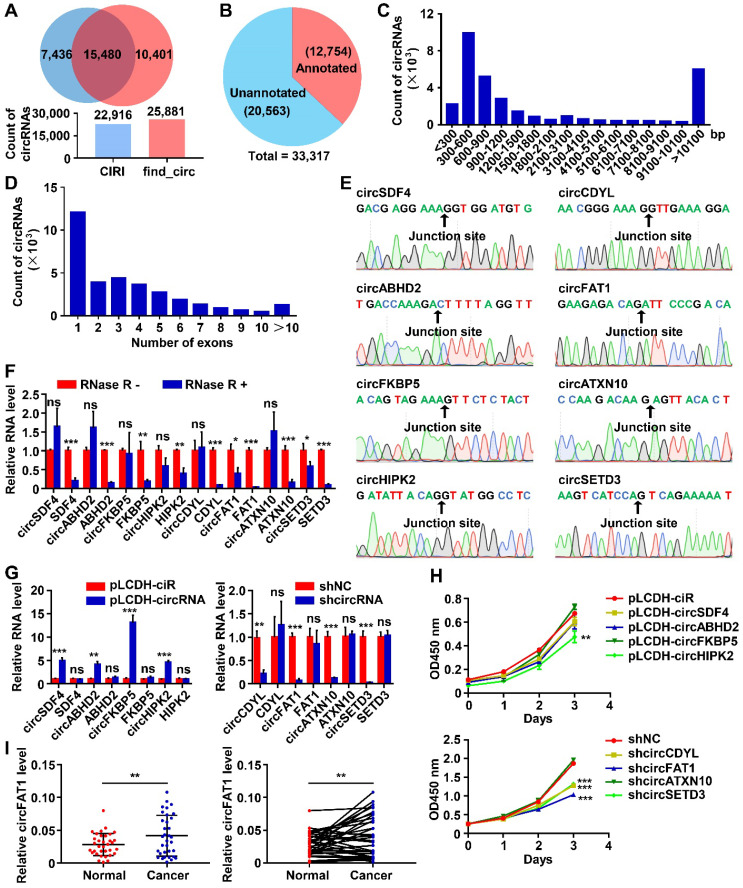
Figure 1.
CircRNA profiling in A549 LUAD cells and functional screening of circRNAs. (A) The numbers of circRNAs identified by CIRI and find_circ from A549 circRNA-seq data. (Upper) Venn diagram showing the numbers of circRNAs identified by CIRI only (light blue), both CIRI and find_circ, and find_circ only (orange); (Lower) The total numbers of circRNA identified by each of the algorithms as indicated. (B) Pie chart showing the numbers of identified circRNAs annotated and unannotated by circBase. (C and D) The distribution of lengths (C) and exon numbers (D) of the identified circRNAs. bp, base pairs. (E) Sanger sequencing confirmed the sequences of 8 circRNAs at backsplicing junction sites. (F) RT-qPCR analyses demonstrating that the indicated circRNAs were more resistant to RNase R digestion than their linear RNA counterparts. (G) RT-qPCR analysis to show the efficiency of circRNA overexpression (Left, pLCDH-circRNAs; pLCDH-ciR for control) or knockdown (Right, shcircRNA; shNC for control) in A549 cells by transducing with corresponding lentiviruses. (H) CCK8 assay to screen circRNAs promoting A549 cell growth. The indicated vectors were used to produce lentiviruses for transducing transgenes into A549 cells. (Upper) circRNA overexpression, (Lower) circRNA knockdown. (I) RT-qPCR assay to analyze relative expression of circFAT1 in paired LUAD tissues and adjacent paracancerous tissues from 34 patients, showing circFAT1 was expressed significantly higher in tumor tissues. GAPDH were used for normalization. *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant.