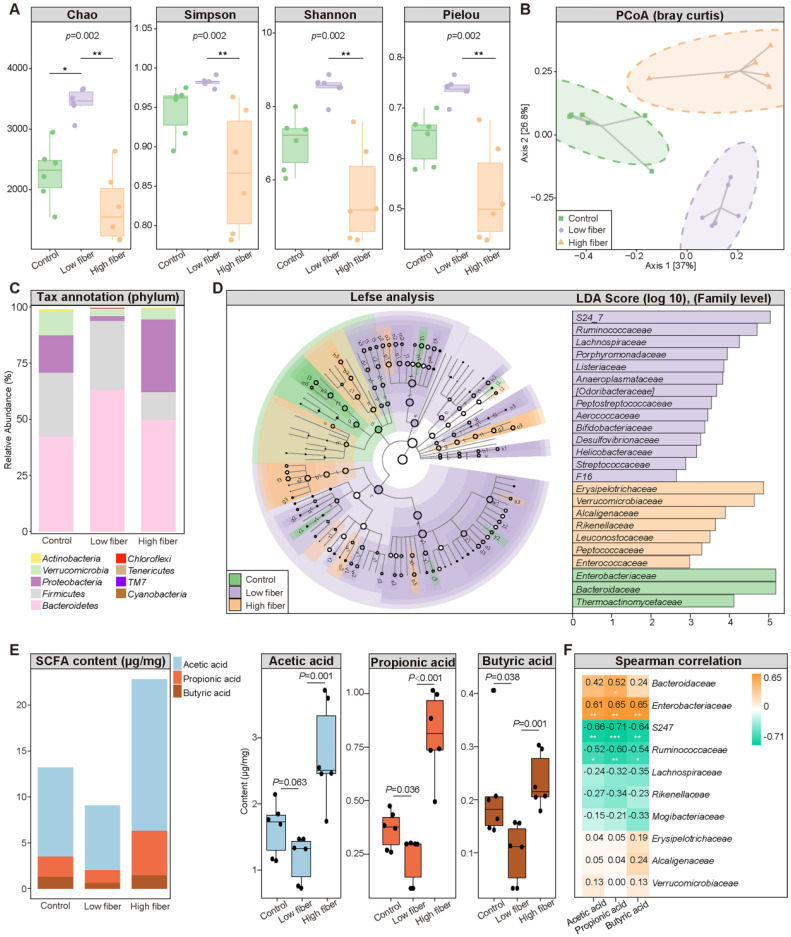
Figure 2.
Dietary fiber alters GM structure and SCFA production. (A) The α-diversity including Chao, Simpson, Shannon and Pielou index in the normal-fiber control (green), low-fiber (purple), and high-fiber (orange) group. Boxes represent the inter quartile ranges, the inside line or points represent the median, and circles are outliers. n=6 for each group, Kruskal-Wallis test. (B) Principal coordinates analysis (PCoA) based on bray curtis distance. The green squares represent normal-fiber control, orange triangles refer to high-fiber, and purple circles denote low-fiber. n=6 for each group. (C) Phylum-level taxonomic abundance and proportion for the normal-fiber control (right), low-fiber (middle), and high-fiber (left) group, where different taxa are differentiated by color. n=6 for each group. (D) Cladogram showing different taxonomic compositions among the normal-fiber control (green), low-fiber(purple), and high-fiber group (orange) based on the linear discriminant analysis (LDA) effect size (LefSe) analysis. Histogram of LDA scores showing differentially abundant taxon (family level). The taxon with |LDA score (log10)| >2 and P < 0.05 are listed, n=6 for each group. (E) Distribution of fecal SCFA levels in control (n=6), low-fiber (n=6), and high-fiber groups (n=6). student's t-test. (F) Spearman's correlation analysis between fecal SCFA levels and the top-10 most abundant genera in high-fiber group. Green, negative correlation; yellow, positive correlation; n=6 for each group; *, P < 0.05; **, P < 0.01, ***, P < 0.001.