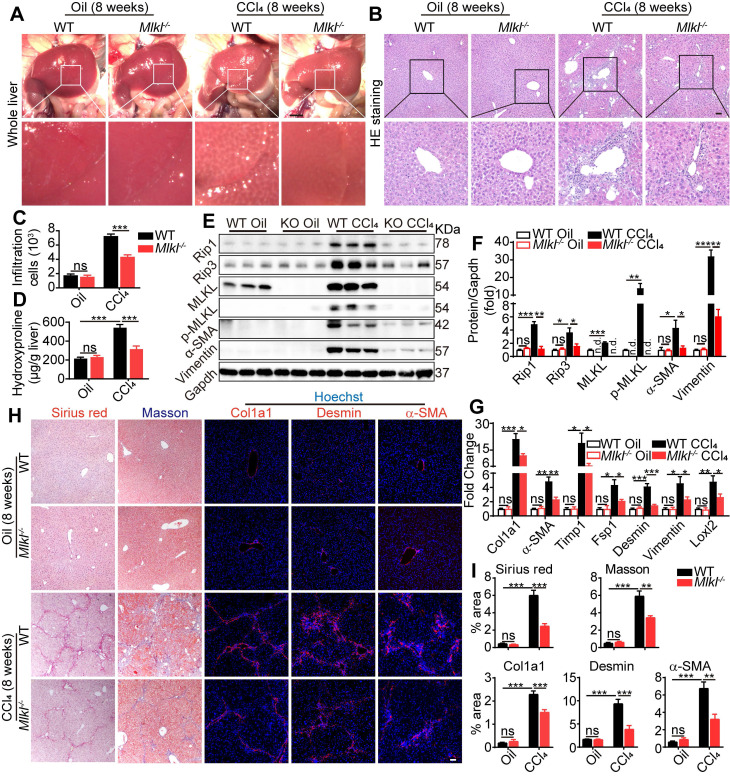
Figure 3.
Deletion of Mlkl attenuates hepatic fibrosis in chronic CCl4 model. (A) Representative whole liver pictures of WT and Mlkl-/- mice treated with oil or CCl4 (chronic). Scale bar represents 0.5 cm. (B) Representative H&E staining images of liver sections. (C) Quantification of the infiltrating cells in (B) (Oil groups, n=4; CCl4 groups, n=8). (D) Hydroxyproline levels in liver samples from WT and Mlkl-/- mice treated with oil or CCl4 (Oil groups, n=4; CCl4 groups, n=8). (E) Western blot analysis of MLKL, p-MLKL, Rip3, Rip1, and fibrosis markers α-SMA, Vimentin in the liver tissues (each lane represents one mouse). Gapdh was used as loading control. (F) Quantification of the blots in (E). All proteins were normalized to Gapdh in the same sample, then normalized to WT mice treated with vehicle (Oil). (G) Quantitative RT-PCR analysis of hepatic fibrosis genes Col1a1, α-SMA, Timp1, Fsp1, Desmin, Vimentin, and Loxl2 in liver samples from WT and Mlkl-/- mice treated with oil or CCl4 (Oil groups, n=4; CCl4 groups, n=8). (H) Representative images of Sirius red and Masson staining, and immunofluorescence staining of Col1a1, Desmin, and α-SMA in liver sections. Nuclei were stained with Hoechst 33342. (I) Quantitative analysis of positive staining areas in (H). (Oil groups, n=4; CCl4 groups, n=8). Data are shown as Means ± SEM, *P < 0.05, **P< 0.01, ***P< 0.001 (Student's t-test). Scale bar represents 100 µm.