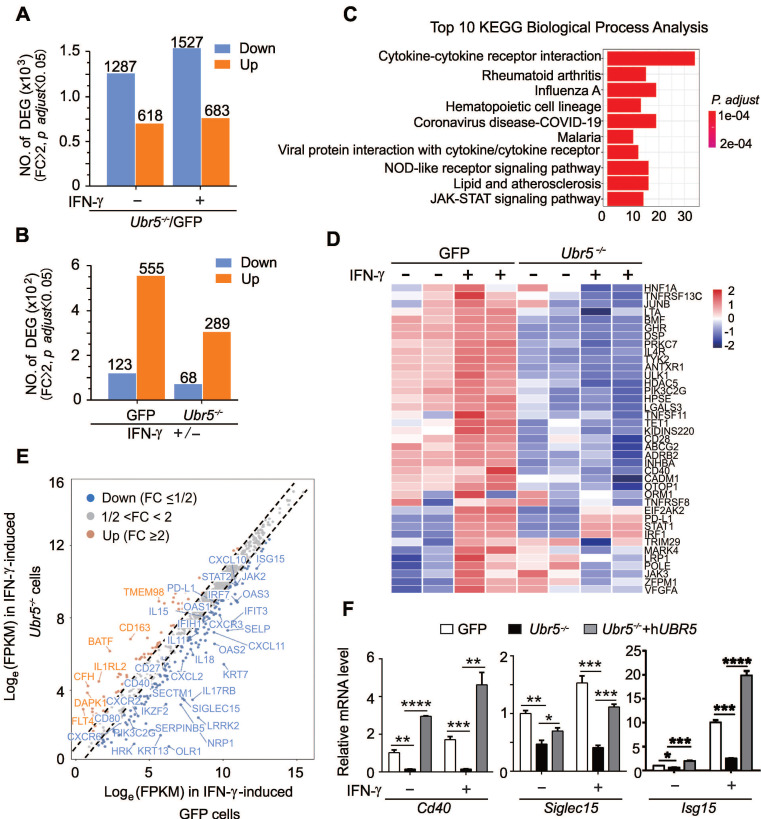
Figure 4.
UBR5 globally regulates IFN-γ-mediated pathways and stimulated genes. (A) Numbers of differentially expressed genes (DEGs) in Ubr5-/- compared with GFP 4T1 cells with or without IFN-γ treatment. Fold change (FC) ≥ 2, p. adjust ˂ 0.05. (B) Numbers of DEGs in GFP 4T1 cells or Ubr5-/- 4T1 cells with IFN-γ treatment compared with that without IFN-γ treatment. Fold change (FC) ≥ 2, p. adjust ˂ 0.05. (C) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment of the top 10 biological process between IFN-γ-treated GFP and Ubr5-/- 4T1 cells. (D) Heatmap depicting the mRNA levels of selected ISGs in GFP and Ubr5-/- 4T1 cells with or without 10 ng/mL IFN-γ induction for 24 h. (E) Scatterplot diagram of genes responding to IFN-γ differently in Ubr5-/- cells compared to that in GFP cells. The grey dots represent unchanged expression (1/2<FC<2) in both cell lines. Upregulated ISGs with fold change (FC) values >2 in Ubr5-/- cells compared with GFP cells are shown with orange dots, while downregulated ISGs with fold change (FC) values <1/2 are shown with blue dots. (F) The mRNA levels of Cd40, Siglec15 and Isg15 were examined by qPCR analysis in WT, Ubr5-/- and hUBR5-reconstituted Ubr5-/- 4T1 cells treated with or without IFN-γ for 24 h. All experiments were repeated three times, and the data are presented as the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.