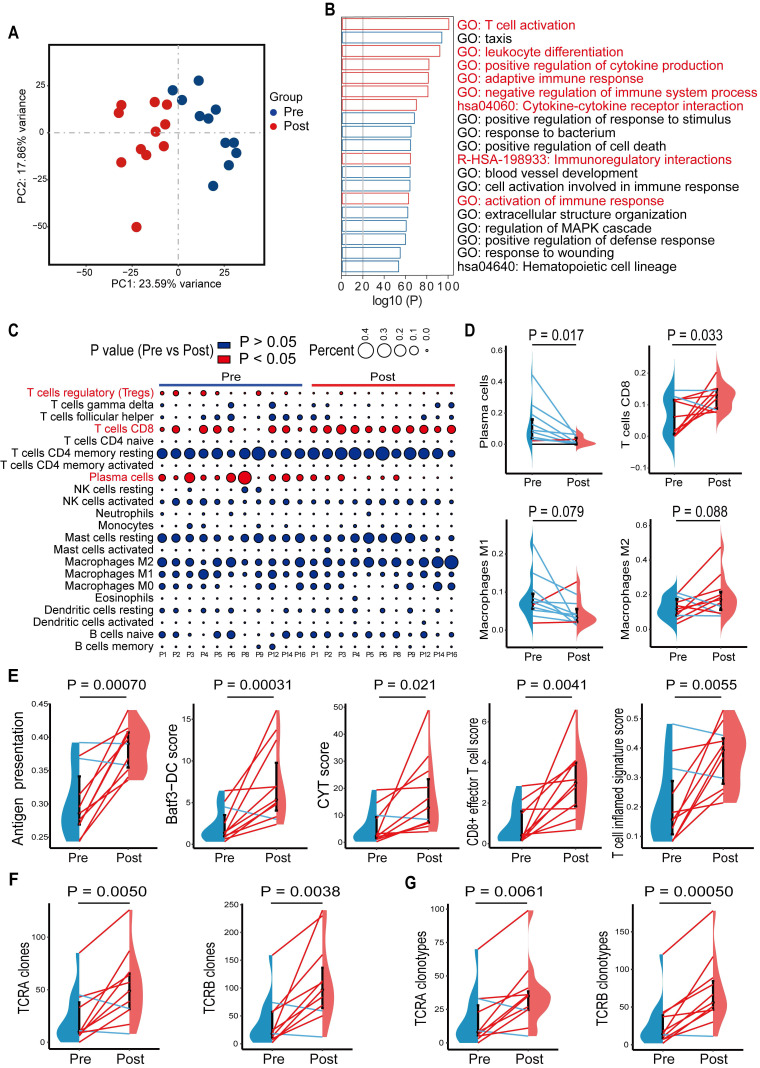
Figure 1.
Gene signatures of tumors before and after chemohormonal therapy in prostate cancer patients. A. Principal component analysis of transcriptomic data from all 22 patient samples. Each dot represents a patient sample that is colored on the basis of treatment (blue, pretreatment; red, posttreatment). B. The horizontal bar graph showing the top 20 of upregulated differentially enriched pathways and functions in post-chemohormonal therapy tumor samples compared to paired pretreatment samples. Red bars indicate the immune-related differentially expressed pathways and functions. C. Bubble plot illustrating relevant immune cell profiles of paired pre- and post-chemohormonal therapy tumor samples. Bubble size reflects the percentage of immune cell subtypes enriched in corresponding immune cell profile. Bubble color reflects the P value. D. Changes in fractions of plasma cells, CD8 T cells, M1 macrophages, and M2 Macrophages between paired pre- and post-chemohormonal therapy tumor samples. E. Changes in antigen presentation score, Batf3-DC signature score, and three T cell-phenotype signatures scores (CYT score, CD8+ effector T cell score, and T cell inflamed signature score) between paired pre- and post-chemohormonal therapy tumor samples. F. Changes in the numbers of TRA and TRB clones detected between paired pre- and post-chemohormonal therapy tumor samples. G. Changes in the numbers of individual TRA and TRB clonotypes between paired pre- and post-chemohormonal therapy tumor samples. Each point represents an independent sample. Data were presented as mean values ± SEM. Paired data were analyzed using the paired t-test or Wilcoxon paired rank test.