Abstract
One-fourth of the global human population is estimated to be infected with strains of the Mycobacterium tuberculosis complex (MTBC), the causative agent of tuberculosis (TB). Using lipidomic approaches, we show that tuberculostearic acid (TSA)-containing phosphatidylinositols (PIs) are molecular markers for infection with clinically relevant MTBC strains and signify bacterial burden. For the most abundant lipid marker, detection limits of ∼102 colony forming units (CFUs) and ∼103 CFUs for bacterial and cell culture systems were determined, respectively. We developed a targeted lipid assay, which can be performed within a day including sample preparation—roughly 30-fold faster than in conventional methods based on bacterial culture. This indirect and culture-free detection approach allowed us to determine pathogen loads in infected murine macrophages, human neutrophils, and murine lung tissue. These marker lipids inferred from mycobacterial PIs were found in higher levels in peripheral blood mononuclear cells of TB patients compared to healthy individuals. Moreover, in a small cohort of drug-susceptible TB patients, elevated levels of these molecular markers were detected at the start of therapy and declined upon successful anti-TB treatment. Thus, the concentration of TSA-containing PIs can be used as a correlate for the mycobacterial burden in experimental models and in vitro systems and may prospectively also provide a clinically relevant tool to monitor TB severity.
Keywords: tuberculosis, MTBC, markers, mycobacteria
Globally, tuberculosis (TB) remains a leading cause of morbidity and mortality with more than 10 million active cases annually.1 The causative agent, Mycobacterium tuberculosis (Mtb), is responsible for approximately 1.5 million deaths per year. Emerging drug resistance such as multidrug-resistant (defined by resistance against rifampicin and isoniazid) TB cases is alarming since treatment fails in a high portion of such patients.2,3 From this perspective, biomarkers that reflect therapy responses are highly desirable to rapidly adjust a treatment to the individual patient’s medical need.4 In this perspective, lipid markers of mycobacterial origin may represent ideal candidates to assess bacterial burden in general.
The cell envelope of Mycobacterium tuberculosis complex (MTBC) bacteria is complex in nature and consists of a plasma membrane (PM) as the innermost layer that is covered by the peptidoglycan (PGN)–arabinogalactan (AG) complex. The AG is further esterified by long-chain mycolic acids (MAs). Into these MAs, various complex lipids and lipoglycans such as sulfolipids (SL), trehalose dimycolates (TDM), phenolic glycolipids, lipoarabinomanan (LAM), and phthiocerol dimycocerosates are noncovalently intercalated, thus forming a strong outer envelope, the so-called mycomembrane (MM).5,6 The mycobacterial PM mainly consists of phospholipids and is similar to those seen in other microorganisms.7 Their main components are phosphatidyl-myo-inositol mannosides (PIM), phosphatidylinositol (PI), phosphatidylethanolamine (PE), phosphatidylglycerol (PG), and cardiolipin (CL). The major fatty acid constituents of the PM are palmitic (16:0), octadecenoic (18:1), and 10-methyloctadecanoic [tuberculostearic acid—TSA, (19:0)]. Mycobacterial lipids represent a pool of potential biomarkers to trace the status of Mtb infections due to their unique structural features not found in eukaryotic lipidomes.8 Due to high diversity and complexity of these lipids, approaches to measure and profile them are challenging.9 Various mass spectrometry (MS)-based lipidomics approaches have been developed during the last 15 years to foster the analysis of lipids, further satisfying the increasing interest on this class of biomolecules.10,11 MAs have been of special interest; however, since standards are ill-defined, the development of MA-based diagnostic markers remains difficult.12,13 The complex lipids of Mtb are comparably large molecules and mostly comprise a neutral net charge, which require specific extraction procedures and ionization and detection conditions.8 In comparison, for phospholipids, numerous and very advanced analytic procedures have been established. This is reflected in a recent study, which showed that different PIM and PI species enable MALDI mass spectrometry imaging (MALDI-MSI) of intact Mtb in rabbit-lung granulomas.14
In the current study, we identified saturated mycobacterial PIs containing TSA as a novel marker of bacterial loads in relevant TB infection models in vitro and in vivo. We further observed that specific PIs were significantly enriched in peripheral blood mononuclear cells (PBMCs) from active TB patients, which may open a future perspective for its use in a clinical setting.
Results
PIs Containing TSA Represent Lipid Markers for Mycobacterial Load
Our search for lipid-based markers of MTBC strains was focused on amphiphilic molecules that should be extractable with established methods, easy to quantify, and present in the pathogen with high abundance. Therefore, we profiled the content of the methanolic fraction by liquid chromatography–mass spectrometry (LC–MS) to perform a targeted analysis of PM lipids (Figure S1). PL profiles of 18 MTBC strains covering five lineages, both ancient and modern ones including three clinical Beijing isolates, exhibited a high similarity (Table 1 and Figure 1). For all investigated MTBC strains we identified, as main molecular species, PE and PI comprising a diacylglycerol (DAG) backbone built from FA 16:0 and FA 19:0 (TSA) (Figure 1A,B). The presence of TSA as the sole fatty acid isomer comprising a 19:0 acyl chain was proven by gas chromatography coupled with mass spectrometry (GC–MS) analysis of the fatty acid methyl ester profiles generated from the methanolic fraction (data not shown), being in line with the literature.15 PE 35:0 (16:0_19:0 (TSA)) is the most abundant molecular species of its class with an average of 31.9 mol %, but its abundance varied to a larger extent between the investigated isolates. PI 35:0 (16:0_19:0 (TSA)) was quantified as major abundant molecular species of its class in all strains with 70.7 mol % on average (Figures 1B,C, S3). It is noteworthy that for the PG class, PG 35:0 (16:0_19:0 (TSA)) was only a minor component, while PG 34:1 was the most abundant species (Figure S2). TSA was further detected in combination with FA 14:0, FA 15:0, FA 16:1, FA 17:0, FA 17:1, and FA 18:0 in PE and PI, as well as LPE 19:0 (lysophosphatidylethanolamine) and LPI 19:0 (lysophosphatidylinositol) in these strains (data file S1). PI 16:0_19:0 (TSA) has the analytical advantage that additional fragments of the PI headgroup can be detected by tandem mass spectrometry (MS2) with high sensitivity and specificity, which is not the case for PE 16:0_19:0 (Figure S4). Thus, PI 16:0_19:0 (TSA) was further studied in detail.
Table 1. Phylogenetic Classification of All Investigated MTBC Strainsa.
| species | strain | lineagea | genotype | origin |
|---|---|---|---|---|
| M. tuberculosis | 12594/02 | 2.2.1 | Beijing | clinical isolate, internal reference collection64,65 |
| M. tuberculosis | 1500/03 | 2.2.1 | Beijing | |
| M. tuberculosis | 1934/03 | 2.2.1 | Beijing | |
| M. tuberculosis | 1797/03 | 1.1.2 | East African Indian | |
| M. tuberculosis | 4850/03 | 1.1.2 | East African Indian | |
| M. tuberculosis | 947/01 | 1.1.3 | East African Indian | |
| M. tuberculosis | 2336/02 | 4.1.2.1 | Haarlem | |
| M. tuberculosis | 4130/02 | 4.1.2.1 | Haarlem | |
| M. tuberculosis | 9532/03 | 4.1.2.1 | Haarlem | |
| M. tuberculosis | 2169/99 | 4.6.1.2 | Uganda | |
| M. tuberculosis | 2191/99 | 4.6.1.1 | Uganda | |
| M. tuberculosis | 2333/99 | 4.6.1.2 | Uganda | |
| M. africanum | 10514/02 | 6 | West African II | |
| M. africanum | 10517/01 | 6 | West African II | |
| M. africanum | 5468/02 | 6 | West African II | |
| M. canettii | 3040/99 | M. canettii | clinical isolate, internal reference collection66 | |
| M. canettii | 3041/99 | M. canettii | ||
| M. canettii | 3151/08 | M. canettii | ||
| M. tuberculosis | H37Rv-ATCC 27294 | 4.9 | American Type Culture Collection, Manassas, USA | |
| M. tuberculosis | H37Rv, mCherry | 4 | described in ref (67) | |
| M. tuberculosis | H37Rv, GFP | 4 | described in ref (53) |
Figure 1.
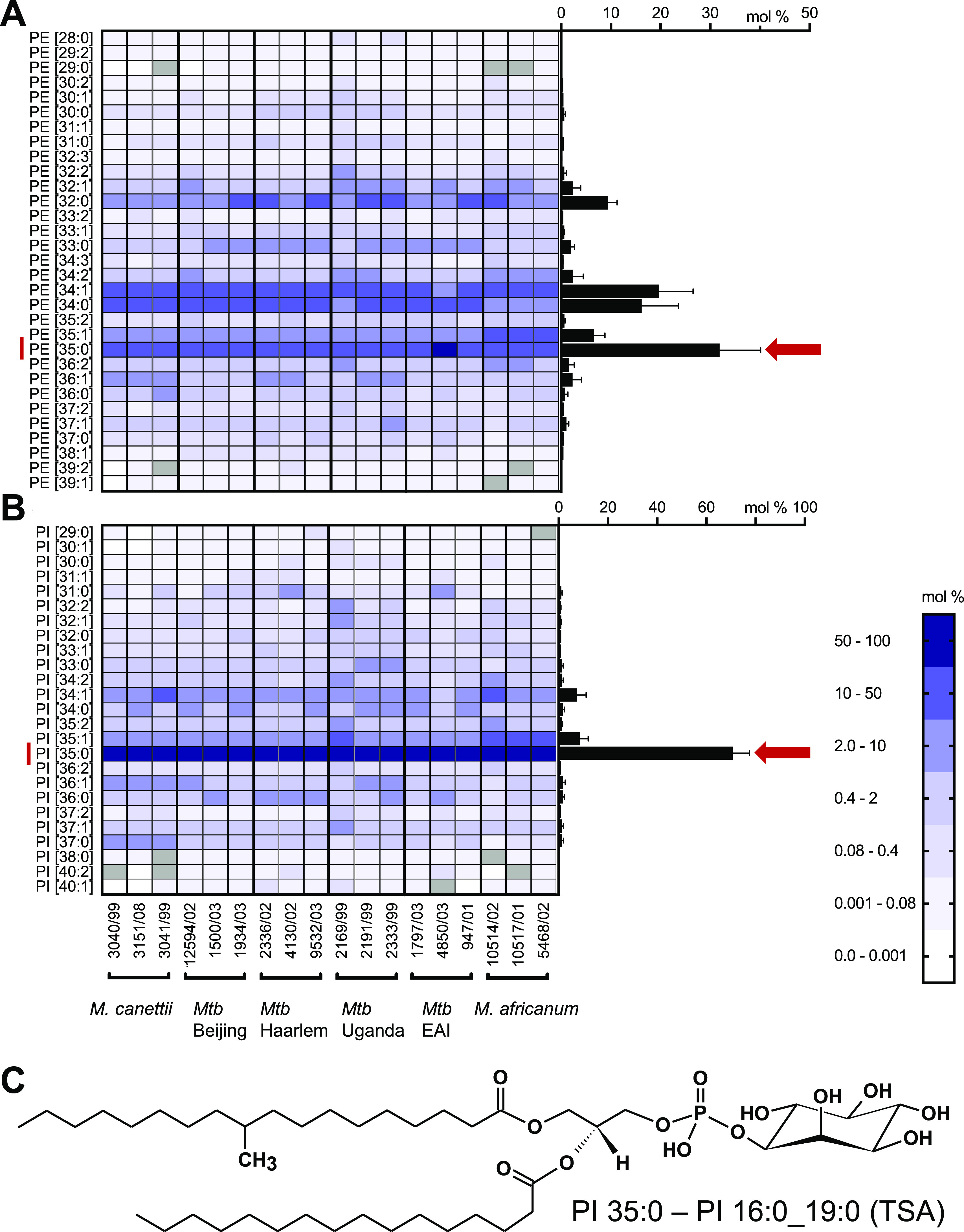
PI 35:0–16:0_19:0 (TSA) is the abundant phosphoglycerolipid of MTBC clinical isolates. Lipid profiles of (A) PE and (B) PI were determined using high-resolution LC–MS from extracts of clinical MTBC isolates. Profiles of clinical isolates were determined from at least two independent cultivations (separate profiles for each strain are shown in Figure S2). On the left, a heat map representation for individual strains is provided. The average lipid profiles for all strains are represented in the right as a bar graph (error bars represent one SD). (C) Structure of the sn-1 isomer of PI 16:0_19:0 (TSA) used as a marker for mycobacterial loads and CFUs. East African Indian strains—EAI.
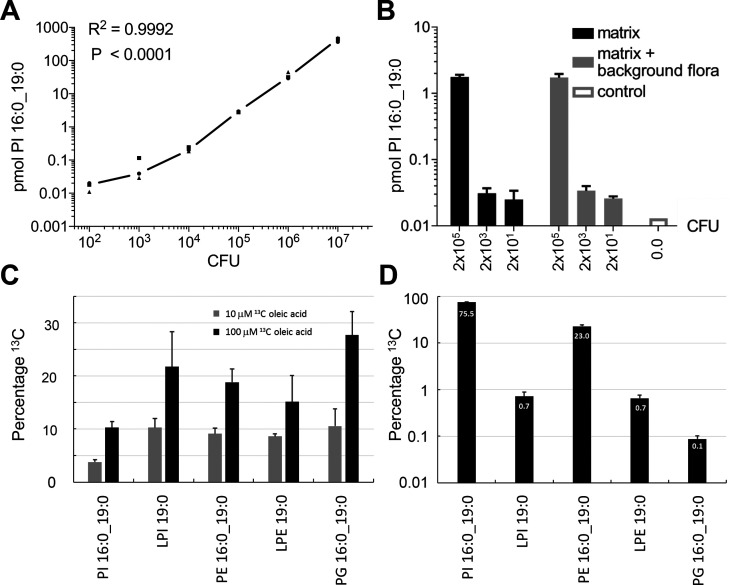
Subsequently, we evaluated whether the PI 16:0_19:0 (TSA) concentration could be used to estimate colony forming units (CFUs) using dilutions of three independent cultures of Mtb H37Rv mCherry. Lipid analysis of bacterial pellets was completed within 24 h including inactivation of Mtb. We observed a linear correlation between the amount of PI 16:0_19:0 (TSA) and CFUs (Figure 2A). Although 100 mycobacteria were still detectable, the lower limit of quantitation for the CFU determination by the PI 16:0_19:0 (TSA) assay was approximately 1000 bacteria. This result indicates that targeted quantitation of PI 16:0_19:0 (TSA) is a fast and sensitive way to determine the CFU of Mtb. From these data, we can estimate that a single mycobacterium contains approximately 45 amol of PI 16:0_19:0 (TSA). This finding is in good agreement with the detection limit of the lipidomics platform for PI molecular species of approximately 1 fmol. The feasibility of this approach was further evaluated in an independent bacteriological test system for sputum analysis. Here, we demonstrated that a mycobacterial load of 2000 bacteria could be detected in both artificial sputum and sputum supplemented with a background flora composed of fungal and bacterial species (Figure 2B).
Figure 2.
Detection of Mtb in culture using PI 16:0_19:0 and metabolic labeling of TSA. (A) Correlation between the amount of PI 16:0_19:0 (TSA) and CFUs (n = 3) of Mtb H37Rv (mCherry). (B) Detection of Mtb (H37Rv) in artificial sputum in the presence of bacterial and fungal flora using PI 16:0_19:0 (TSA). (C) Labeling efficiency of Mtb using 13C-labeled OA (FA 18:1, OA) which is metabolized to FA 19:0 (TSA) by the pathogen and (D) relative abundance of main FA 19:0 (TSA)-containing membrane lipids.
Tracer Analysis Using 13C-Labeled Oleic Acid and Its Transformation to TSA Enables Monitoring of Mtb Growth
The biosynthesis of the PM is crucial for Mtb growth. Therefore, a tracer analysis of its biosynthesis will provide a well-defined estimate for mycobacterial replication. Oleic acid (OA; FA 18:1) was chosen as the substrate molecule, a highly abundant building block of membrane lipids and neutral lipids in mammalian cells. When the growth medium was supplemented with 13C18-labeled OA (13C18 OA), we observed its transformation to TSA by the addition of a methyl group comprising one 12C atom (Figure S5–S8). Such labeled TSA served as a building block for the membrane lipids PI 16:0_19:0 (TSA), PE 16:0_19:0 (TSA), and PG 16:0_19:0 (TSA) and LPI 19:0 (TSA) and LPE 19:0 (TSA) (Figures 2C, S5–S8). After 7 days of incubation, we observed dose-dependent labeling efficiencies for PI 16:0_19:0 (TSA) of 3.8% with a supplement of 10 μM 13C18 OA, which was further increased to 10.3% for 100 μM for the tracer molecules. The labeling efficiency for LPI 19:0 (TSA), PE 16:0_19:0 (TSA), LPE 19:0 (TSA), and PG 16:0_19:0 (TSA) was approximately doubled for both conditions when compared to PI 16:0_19:0 (TSA), but in terms of total amounts, PI 16:0_19:0 (TSA) was the major labeled species (Figure 2D). An even higher labeling efficiency for PI 16:0_19:0 was recently observed when Mtb was cultured together with macrophages previously labeled with 13C18 OA.16
Quantitation of PI 16:0_19:0 Enables Monitoring of the Mtb Infection Status in Cell Culture Systems
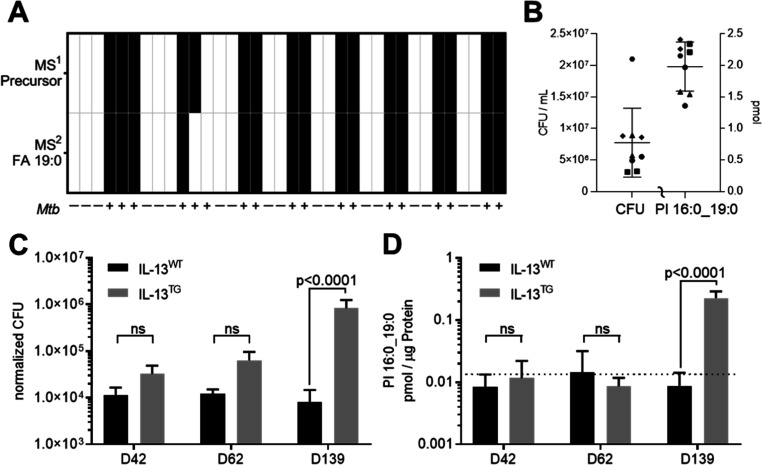
Next, we used MS2 to perform a targeted analysis of PI 16:0_19:0 (TSA) to monitor all isomers in the mass range of 851.5 ± 0.5 Da (Figure S4) in order to address the infection status and the mycobacterial load in cell culture systems. For 36 independent cultures of murine bone marrow-derived macrophages (BMDMs), the infection status was correctly determined with only one false negative test (Figure 3A). The Mtb infection status was determined using two mass spectrometric signals, the precursor ion at m/z 851.5658 and the FA 19:0 (TSA) fragment ion 297.2798 (Figure S5A). The applicability of PI 16:0_19:0 as a marker of mycobacterial load was further analyzed in infection experiments with human neutrophils. Neutrophils from five healthy donors were infected with a multiplicity of infection (MOI) of 3 and incubated for 6 h. After this short incubation time, phagocytosis is completed but intracellular pathogens have not yet multiplied nor were the mycobacteria killed by the neutrophils.17 This approach enabled us to evaluate if a basal concentration of PI 16:0_19:0 (TSA) was present in human neutrophils. All control cultures were identified as negative (n = 11), and all cultures infected with Mtb were identified as positive (ion at m/z 851.5658 in MS1 and fragment ion 297.2798 in MS2 detectable; n = 9) (Figure 3B). The experimental variation for this standardized infection model was further compared by relative standard deviation (RSD). Lipid quantitation had a better precision with an RSD of 0.20 when compared to the CFU determination with an RSD of 0.70 (Figure 3B). These results indicated that PI 16:0_19:0 (TSA) is specific enough to detect and quantify Mtb load in microbiological experiments and murine and human cell culture models.
Figure 3.
Application of PI 16:0_19:0 as a correlate of Mtb load in model systems. (A) Detection of Mtb in murine BMDM. MS detection level either on basis of accurate mass determination in MS1 (m/z 851.5655) or MS2 (FA 19:0, m/z 851.5 → 297.2799; Figure S4A) (B) validation of the Mtb infection status for neutrophils isolated from five healthy donors. At 6 hpi, CFUs (left axis) and amount of PI 16:0_19:0 (right axis) were determined. Data from different donors are indicated by distinctive symbols with double determination for each individual. In noninfected cultures, PI 16:0_19:0 was below the limit of detection. (C) Pulmonary mycobacterial burden in IL-13WT and IL-13TG mice upon Mtb aerosol infection by CFU analysis at indicated time points and (D) respective concentrations of PI 16:0_19:0 determined from the same lung tissue homogenates. The dotted line indicates basal concentration in uninfected mice (1.75 × 10–2 pmol/μg; n = 11, data file S2). One-way ANOVA analysis was performed for data presented in panels (C,D).
Monitoring CFUs in Mtb-Infected Mice by Quantitation of PI 16:0_19:0
Many studies on fundamental processes during Mtb infection are performed in experimental mouse models. Of particular interest are models which reflect TB disease in humans.18,19 This holds also true for IL-13TG animals, in which Mtb infection—in contrast to IL-13WT (BALB/c) mice—leads to unrestricted bacterial growth and subsequent development of centrally necrotizing granulomas, which strongly resemble the morphology of lesions in TB patients.20,21 In the present study, we monitored the growth of Mtb in IL-13TG and IL-13WT mice as well as CFU development and the presence of PI 16:0_19:0 at day 42, 62, and 139 post infection (p.i.). We observed a significant increase in bacterial numbers in lungs of IL-13TG when compared to IL-13WT mice 139 days after infection (Figure 3C). In Mtb-infected IL-13WT control mice, CFU levels measured at d42, d62, and d139 stayed at approximately 1.0 × 104 bacteria per extracted tissue sample (4.7 × 106 CFU/g tissue, data file S2) during Mtb infection, which corroborates previous studies that CFU levels in the lung of C57Bl/6 mice remain rather stable during chronic experimental TB.22 We next determined PI 16:0_19:0 levels in lung homogenates of these mice. During the whole course of infection, PI 16:0_19:0 concentrations stayed at ∼1.1 × 10–2 pmol/μg in IL-13WT mice (Figure 3D), a value which was below the level of uninfected mice of 1.75 × 10–2 pmol/μg (n = 11, dotted line in Figure 3D), which may indicate a metabolic background obtained from mice housed in an animal facility. It should be noted that for in vivo samples, the mass spectrometry-detected 19:0 FA comprises besides TSA also other isomers representing a metabolic background from microbiota and nutritional sources (Figure S4). Levels of PI 16:0_19:0 in lung homogenates of IL-13TG were comparable to those in IL-13WT mice during the early time points of the infection (d42, d62 p.i.); however, at d139 in parallel with a significant increase in bacterial burden, the amount of PI 16:0_19:0 increased and correlated with the Mtb loads in tissue samples (Figure 3D and data file S2). Taken together, the detection limit at which PI 16:0_19:0 in tissue samples can be specifically recognized beyond the metabolic background of 1.75 × 10–2 pmol/μg was estimated to be 1.0 × 105 bacteria for an extracted lung tissue homogenate (Figure 3D). Of note, whereas classical CFU determination requires 3–4 weeks before growth of Mtb could be determined, targeted quantification of PI 16:0_19:0 enables us to quantify increased Mtb loads in susceptible mice within 24 h. However, the threshold of PI 16:0_19:0 concentration as a correlate of CFUs is only applicable for mouse models susceptible to Mtb infection, where bacterial loads are relatively high.
Quantitation of PI 16:0_19:0 and Putative Transesterification Products in PBMCs from TB Patients and Healthy Controls
Next, we investigated whether the detection of PI 16:0_19:0 could be of potential use in a clinical setting. Attempts to monitor PI 16:0_19:0 levels in plasma samples from healthy controls and TB patients were not successful, very likely due to differences in the sampling time that led to differences in circadian rhythm and nutritional status.23,24 We then hypothesized that mycobacterial PI 16:0_19:0 is detectable in PBMCs isolated from active TB patients as these cells may have taken up this molecule as they are constantly migrating in and out of granulomatous lesions during the immune response against the pathogen.25,26
First, we analyzed PBMCs of 39 healthy controls and determined a metabolic background in these samples of about 0.13 pmol/1 × 106 cells (median, n = 38, data file S3), which for one individual PI 16:0_19:0 was below the detection limit. The same analysis of PBMCs from TB patients at therapy start (t0) showed significantly increased PI 16:0_19:0 levels of 0.3 pmol/1 × 106 cells (median, n = 23) when compared to healthy controls (Figure 4A and data file S4). Receiver operating characteristic were determined in comparison to the healthy controls with an AUC = 0.784 and a sensitivity of 69.7% and a specificity of 79.0% at the optimal cutoff value (Table S1 and Figure S9). From the cohort of TB patients, we then focused on the subset of patients who either completed anti-TB therapy (WHO outcome criteria, Figure 4B, n = 13) or were cured according to the TBnet outcome criteria,2 which involves a follow up of 1 year after therapy end (Figure 4C, n = 9). We compared the levels of PI 16:0_19:0 in PBMCs at t0 and at the end of therapy (tE, Figure 4C and data file S5). Considering the WHO criteria, PI 16:0_19:0 levels decreased in 9/13 patients, stayed similar in 3/13 patients, and increased in 1/13 patients when t0 levels were compared toward tE. With regard to TBnet criteria, PI 16:0_19:0 levels decreased for 7/9 and remained similar for 2/9 patients (Figure 4B,C). Taken together, with both outcome definitions, we observed that PI 16:0_19:0 levels in PBMCs of these patients were significantly reduced at the end of therapy when compared to therapy naive patients.
Figure 4.
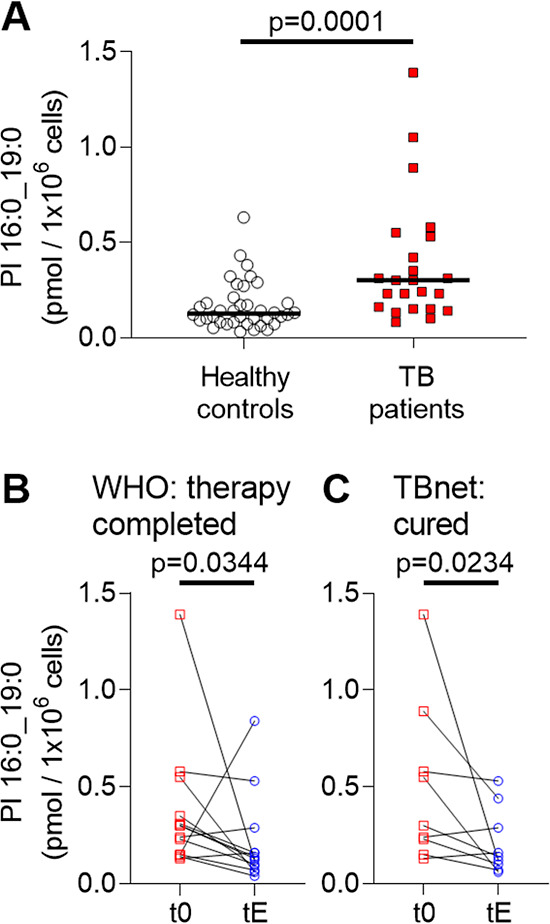
Amount of PI 16:0_19:0 in PBMCs of TB patients are indicative of disease status. (A) Concentration of PI 16:0_19:0 in PBMCs of 39 healthy individuals (n = 1 negative for PI 16:0_19:0) and 23 TB patients at therapy start (t0). The median is indicated with a black bar. Statistical significance was examined with the Mann–Whitney test (individual data from patients and healthy controls are listed in data files S3 and S4). (B) PI 16:0_19:0 concentration at therapy start (t0) and at therapy end (tE) for 13 patients with the WHO-defined outcome “therapy completed”. (C) Comparison of PI 16:0_19:0 concentration for nine patients at therapy start (t0) and therapy end (tE) that further fulfilled the outcome criteria “cured” following the TBnet criteria. Statistical examination for (B,C) was performed with a Wilcoxon matched-pair signed rank test. All statistical tests were performed with GraphPad Prism 8.4.3. Data for comparison of TB patients at t0 and tE according to the outcome criteria definition are listed in data file S5.
We further hypothesized that host cells may have metabolized a certain amount of mycobacterial PI 16:0_19:0 via transesterification to form host cell lipids that contain TSA. Specifically, PI 19:0_20:4 caught our interest since it was detected with up to 20-fold higher amounts in PBMCs when compared to PI 16:0_19:0 and showed even slightly improved accuracy (Figure S10 and Table S1). This molecule is structurally very similar to the major abundant endogenous species PI 18:0_20:4 and might be produced with high transesterification rates. We further included seven additional PI species comprising FA 19:0 that were regularly detected in PBMCs into a marker panel (sum FA 19:0, Table S2). For PBMC samples, the marker panel comprises a mix of PI isomers (Figure S4) where we expect that for TB patients increased contribution from TSA-containing PIs is quantifiable. A comparison of the overall amount of this marker panel between healthy controls and therapy naive TB patients showed such a significant increase from a median of 5.1 pmol/1 × 106 to 13.2 pmol/1 × 106 cells, respectively (Figure 5A and data files S3 and S4). Furthermore, the accuracy for the comparison of TB patients and healthy controls was slightly better than for PI 16:0_19:0 alone with an AUC = 0.802, a sensitivity of 73.9%, and a specificity of 82.1% (Table S1 and Figure S11). We then again focused on patients that were grouped either according to the WHO or the TBnet outcome definitions. The concentration levels of all FA 19:0-containing PIs in PBMCs showed a significant reduction at tE when compared with t0 (Figure 5B,C and data file S5). In 9 out of 10 patients who were cured according to the TBnet definition, the concentration of sum FA 19:0 was lower in PBMCs at tE when compared to t0 (Figure 5C).
Figure 5.
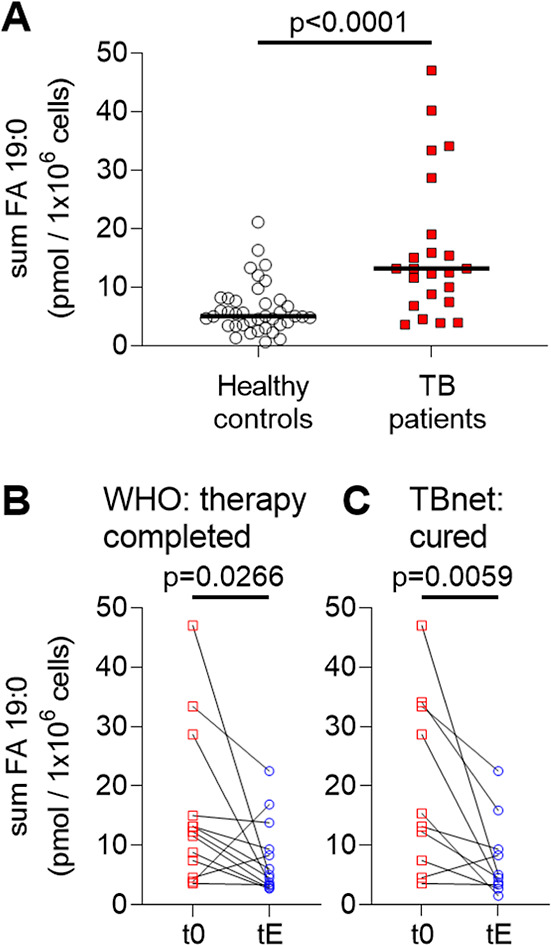
FA 19:0-containing PIs in PBMC correlate with therapy outcome for TB patients. (A) Comparison of summed amounts of 9 FA 19:0-containing PIs in PBMCs (sum FA 19:0; Table S2) of 39 healthy individuals with 23 TB patients at therapy start (t0). Median values are indicated with a black bar. Statistical significance was examined with the Mann–Whitney test (data files S3 and S4). (B) Amounts of sum FA 19:0 at the therapy start (t0) and at therapy end (tE) for 13 patients with the WHO-defined outcome “therapy completed”. (C) Comparison of sum FA 19:0 for 10 patients at therapy start (t0) and therapy end (tE) that further fulfilled the outcome criteria “cured” following the TBnet criteria. Statistical examination for (B,C) was performed with a Wilcoxon matched-pairs signed rank test. All statistical tests were performed with GraphPad Prism 8.4.3. All data for paired statistical analyses according to TB outcome criteria are listed in data file S5.
Discussion
We detected PI 16:0_19:0 (TSA) in 18 clinical MTBC isolates as abundant mycobacterial determinant representing different phylogenetic lineages. PI 16:0_19:0 quantifications correlated with CFUs in broth cultures, cultures from murine macrophages and human neutrophiles, and tissue samples of experimentally Mtb-infected mice with a susceptible genetic background. Measuring PI 16:0_19:0 concentrations allowed us to rapidly determine the mycobacterial burden within 1 day, while CFU-based enumeration requires up to 2 months. We further showed that biosynthesis of TSA by the pathogen and its incorporation into not only PIs but also other phosphoglycerolipids can become an interesting tool for metabolic studies (Figure S12A). Furthermore, the first proof-of-principle experiments using PBMCs from active TB patients showed enhanced levels of PI 16:0_19:0 and FA 19:0-containing PIs when compared to healthy controls and cured TB patients. We envisage that our study on the metabolic interaction of Mtb and its host, with focus on major abundant PIs containing FA 19:0, might encourage follow-up studies in the TB community (Figure S12B). In the following, we discuss the open questions for the application of this lipid panel in translational research and clinical settings.
PI 16:0_19:0 (TSA) as a Marker for Mtb CFUs and Metabolic Activity
In this study, we demonstrate that PI 16:0_19:0 can be used as a molecular marker of Mtb quantification in the background of complex sample matrices of mammalian origin. The direct link of PI 16:0_19:0 to the composition of the PM provides a solid basis for quantification of mycobacterial loads across the MTBC. Quantification of PIs is well established in lipidomics, and appropriate internal standards for absolute quantitation are available.27 In addition, the analytical sensitivity for Mtb detection can be improved by targeted analysis using modern triple quadrupole instruments in combination with LC. This could improve detection limits that were in the current study approximately 100 CFUs in bacterial culture systems and around 1000 bacteria in cell culture systems. Tracer analysis using isotopically labeled OA will enable studying general aspects of the Mtb lipid metabolism. The incorporation of one 12C methyl group by Mtb to form 12C113C18-19:0 (TSA) provides a very specific mass spectrometric tag to perform pulse-chase-type experiments in basic TB research. The enzymatic transformation of FA 18:1 (OA) to FA 19:0 (TSA) has been controversially discussed, and two mechanisms have been proposed. Initially thought to be solely associated with Rv0447c, a two-step mechanism was suggested for Mycobacterium chlorophenolicum, putatively catalyzed by proteins WP_048472120 and WP_048472121, orthologues of WP_003917236 and WP_003420415 (corresponding to Rv3719 and Rv3720 of H37Rv).28,29 We are convinced that further detailed analyses of the enzymatic transformation of FA 18:1 (OA) to FA 19:0 (TSA) in appropriate model systems will support studies on Mtb metabolism with regard to replication, dormancy, persistence, and also reactivation.
Our understanding of the metabolization of PI 16:0_19:0 and TSA during interaction with the host into the metabolites described herein is incomplete. Most prominently, PI 19:0_20:4 as a surrogate mycobacterial marker suggests putative metabolization routes in the host for PI 16:0_19:0 (TSA) and/or other TSA-containing lipids via transesterification. However, basal levels for PI 19:0_20:4 and other FA 19:0-containing PIs were detected in noninfected murine lung tissues (data file S2), human neutrophils, and PBMCs of healthy controls. This result indicates that interferences through nutritional and/or microbial factors need to be considered in future analyses. Finally, the metabolic steps responsible for the transformation of mycobacterial PI 16:0_19:0 or FA 19:0 (TSA)-containing lipids to PI 19:0_20:4, most likely linked to salvage pathways from lysosomes and phagosomal processing, are yet to be discovered (Figure S12).
Quantifying Mycobacterial-Derived Metabolites
Mycobacteria-derived (glyco-)lipid markers such as MAs,12,13 LAM,30,31 and also TSA32−37 were already introduced for the diagnosis of active TB. Specifically, LAM assays were developed to identify TB cases in resource-limited settings. MAs were applied as the discriminator in the clinical diagnosis of TB and might be used in drug efficacy analyses in mouse models.12 However, we assessed it critical that a fixed stoichiometry per bacteria cannot be expected for MAs as the building block of the highly lipophilic outer membrane layer of the mycobacterial cell wall. MAs are not only mainly conjugated to the PGN–AG complex but also covalently linked to trehalose to form TDM and trehalose MM and further present as free MAs.6,38,39 Therefore, MAs represent a dynamic class of cell wall molecules that cannot be assumed to be conserved in quantity, structure, and composition during host–pathogen interactions since they originate from several characteristic cell wall components that contain hundreds of molecular species. Specifically, TDM is expected to be generated in different quantities per bacteria as it is a virulence factor involved in hindering phagolysosome fusion.40−42 Indeed, it was demonstrated that the amounts of TDM are enhanced in mycobacteria during the intracellular life phase in macrophages.43 While the complex regulation of MAs by Mtb is of utmost interest for the basic understanding of host–pathogen interaction, we found that the degree of structural complexity, especially caused by various modifications such as cyclopropane rings, methyl branches, ketones, and methoxy groups,38,44 and unknown metabolic pathways in the host are discouraging for further marker development. Even more important, it was shown that the MA profiles are strain-dependent,12 and we aimed to utilize marker molecules that are present in the entire MTBC.
FA 19:0 (TSA) as a marker has two major advantages: it is present in large amounts in all prototype isolates of the MTBC we investigated so far and its quantitation can be performed by GC–MS.37,45 However, as a chemical building block, it is present in a number of important mycobacterial lipids such as the major abundant PL classes PE and PI and PIMs,46 LM, and LAM.47 Specifically, the latter complex glycoconjugates are reported as virulence factors. Glycolipids are either inserted in the PM stretching through the cell wall to the outer surface or are linked to the outer membrane.48 In analogy to MAs, it is likely that the amount of FA 19:0 (TSA) per bacteria varies depending on environmental conditions, metabolic status, and host–pathogen interaction. In contrast, the PI composition of the PM was well conserved between all investigated MTBC strains. Thus, our focus on FA 19:0 (TSA)-containing PIs and the major abundant species present in the PM of Mtb could provide a robust correlation to the mycobacterial burden in experimental model systems (Figure S12A).
Perspectives for Mycobacterial-Derived Lipid Biomarkers in Clinical Application
Commercially available PCR-based test systems such as the GeneXpert MTB/RIF ultra (Cepheid, Sunnyvale, Ca., USA) are very sensitive with regard to the detection of Mtb but are most probably not suitable for therapy response assessment due to extended mycobacterial DNA stability even after the death of the bacteria.49,50 However, PCR-based methods are best suited to predict drug resistance for most anti-TB agents and genetic classification of strains, which is not possible based on the described PI lipid panel.51 Currently, culture remains the gold standard for the diagnosis of TB and the assessment of bacterial burden despite being slow. In the present study, we performed first analyses with regard to the presence of PI 16:0_19:0 and/or other FA 19:0-containing lipids in TB patient PBMCs. In a future perspective, we nevertheless could envisage that quantitation of PI 16:0_19:0 may allow a more rapid estimation for Mtb burden even during TB therapy, which could be potentially expanded to sputum samples as well. In order to get to this point, detailed longitudinal studies of patient samples taken at different time points of therapy are required to foster such an approach (Figure S12B).
There is ample evidence for the strong dependency of the plasma lipidome composition on the sampling time caused by circadian rhythm and nutritional status.23,24 In addition, several factors related to the patient’s health and to local healthcare settings may prevent a timely blood sampling at the same time point. This is why we focused our analytical developments on the quantitation of lipid markers in PBMCs. Using this strategy, we aimed to minimize the variation in lipid composition caused by the metabolic state of patients. With regard to this notion, our findings are encouraging to further explore mycobacterial lipid biomarkers in clinical application based on well-established PBMC isolation protocols. However, more data are required to establish a causality between mycobacterial burden and the disease state of a patient with the quantity of detectable TSA-containing PIs. In our study, we saw a limit in the realistic detection limit of TSA-containing PIs, which can be attributed to losses during sample preparation procedures. There is still some potential to improve the direct application of the sensitivity of mass spectrometric measurements in the amol range by streamlining chemical extraction procedures.
In the presented study, we did not address in detail whether an interaction between response profiles of FA 19:0-containing PIs and PBMC cell populations exists. Further studies are required to evaluate such correlations and whether performance of the marker could be improved by focusing on specific subsets of immune cells such as professional phagocytes. We are confident that the presented approach for the clinical sample preparation could also be performed in low-resource countries when sample storage and transport would follow general sample handling guidelines. Specifically, an early addition of methanol and the antioxidant butylhydroxytoluol (BHT) to PBMCs should sufficiently block inherent enzymatic activities and oxidation. This can be prepared as sampling kits that would be used at point of care. As diagnostic markers, PI molecules are chemically stable enough to establish recruitment and sampling strategies, which afterward make it a logistical question how the transfer of patient samples to dedicated MS facilities is realized.
Methods
Mycobacteria
MTBC strains (Table 1) were grown in Middlebrook 7H9 broth (Difco, Detroit, USA) supplemented with 10% Middlebrook OA albumin-dextrose-catalase (OADC) enrichment medium (Life Technologies, Gaithersburg, USA), 0.2% glycerol, and 0.05% Tween 80 in 490 cm2 Corning roller bottles. Cultures were harvested at mid log phase (OD600 = 0.3–0.6), aliquoted, and frozen at −80 °C.
Cultivation and Lipidomics Analysis of MTBC Clinical Isolates
Frozen aliquots (OD600 = 0.3–0.6) of clinical isolates were thawed and homogenized by transferring bacterial suspension through a 1 mL syringe. A total of 300–500 μL of bacterial culture was used for inoculation in a final volume of 10 mL. Cultures were incubated at 37 °C and 5% CO2 for 7–10 days. Contamination controls on blood agar and Ziehl–Neelsen staining were performed repeatedly. The bacterial load was determined by CFU analysis on 7H10 agar plates containing 10% bovine serum and 0.5% glycerol. The analytical strategy to profile mycobacterial lipids is summarized in the Supporting Information (Figure S13). Briefly, bacterial pellets of 5.0 × 108 bacteria were transferred into glass tubes containing 2 mL of petroleum ether (bp 60–80 °C) and 4 mL of methanol for inactivation of bacteria. Only the methanolic phase was analyzed with LC–MS using a 1100 HPLC system (Agilent, Waldbronn, Germany) coupled to an Orbitrap Q Exactive Plus (Thermo Fisher Scientific, Bremen, Germany). The methanolic phase (100 μL) was dried under vacuum and reconstituted in 100 μL of CHCl3/MeOH containing 0.1% ammonium acetate 86/13 (v/v), and 5 μL was used for injection. Separation of lipids was performed on a 150 mm BETASIL Diol-100 column with a particle size of 5 μm and 0.32 mm inner diameter (Thermo Fisher Scientific, Bremen, Germany). Detailed information for LC–MS data acquisition is given in the Supporting Information.
Detection of PI 16:0_19:0 as Surrogate for CFU Determination
Aliquots of mCherry expressing Mtb H37Rv (OD600 = 0.4) were thawed and centrifuged (4200g, 10 min, 4 °C). After resuspending the pellet in PBS, defined amounts ranging from 1.0 × 102 to 1.0 × 107 bacteria were transferred into tubes and again centrifuged (4200g, 10 min, 4 °C). Pelleted bacteria were then agitated in 400 μL of methanol for 16 h prior to lipid extraction and shotgun lipidomics analysis.
Detection of Mtb in Artificial Sputum with Targeted Analysis of PI 16:0_19:0
Artificial sputum was prepared according to a customized protocol52 and spiked with the known CFU of Mtb H37Rv and background flora composed of fungal and bacterial pellets. Pellets were mixed with 1 mL of MeOH to kill bacteria in the biosafety laboratory level 3 and transferred to the chemical laboratory. The samples were dried down in a SpeedVac vacuum concentrator (Thermo Fisher Scientific, Waltham, US) before they were resuspended in 50 μL of water. The suspension was thoroughly mixed and shock-frozen in liquid nitrogen. Afterwards, samples were transferred into an ultrasonic bath (Sonorex, Bandelin, Berlin, Germany) filled with ice-cooled water. Samples were sonicated for 1–2 min until thawing was complete. This freeze/thaw cycle was repeated three times. Then, the modified methyl-tert-butyl ether (MTBE) protocol described below was performed. The organic phase was dried down and dissolved directly in 100 μL of MS electrospray ionization solution consisting of CHCl3/MeOH/isopropanol 1/2/4 (v/v/v) with 0.05 mM ammonium chloride for shotgun lipidomics analysis.
Isotopic Labeling of Mtb Lipids
2.0 × 106 GFP-expressing Mtb bacteria H37Rv were cultured in a microtiter-based format with a volume of 100 μL50 in 7H9 medium containing 0.5% bovine serum albumin, 0.085% NaCl, 0.0003% catalase, and 0.04% Na-acetate. Culture media were supplemented with natural isotope distribution (12C) or completely 13C-labeled OA (12C: O1383, Sigma-Aldrich, Taufkirchen, Germany; 13C: CLM-460-0 (98%), Cambridge Isotope Laboratories, Tewksbury, US). After 7 days of culturing, bacterial suspensions were centrifuged at 4200g for 10 min at 4 °C, followed by removal of the supernatants. The pellet was resuspended and agitated in 500 μL of methanol for 2 h at RT prior to lipid extraction and shotgun lipidomics.
Generation of BMDMs and Infection with Mtb
Murine BMDMs were isolated as described earlier (details are given in the Supporting Information).54,55 For infection of BMDMs, frozen aliquots of Mtb H37Rv were thawed and centrifuged (2300g, 10 min), and bacteria in cell culture medium were carefully homogenized using a 26-gauge syringe needle. Subsequently, cells were infected with an MOI of 1 and were incubated for 4 h (37 °C, 5% CO2), followed by extensive washing with Hanks’ buffered salt solution (Sigma-Aldrich, Taufkirchen, Germany) in order to remove extracellular bacteria. Cells were cultured for a total of 72 h as previously described.56 Afterward, supernatants were discarded, and cells were washed once with PBS (37 °C). Subsequently, 100 μL of ice-cold water was added, and cells were incubated on ice for 30 min. Cells were detached using a cell scraper and transferred into suitable tubes (Eppendorf, Hamburg, Germany) containing 270 μL of MeOH (hypergrade for liquid chromatography, Merck, Darmstadt, Germany).
Isolation and Infection of Peripheral Human Neutrophils
Human neutrophils were isolated from peripheral blood of healthy volunteers (two female and three male donors of normal weight) aged 30–40 years. For the isolation of peripheral neutrophils, granulocytes were purified by density centrifugation for 25 min at RT at 800g through a Histopaque 1199 gradient (Sigma Aldrich, Taufkirchen, Germany). A discontinuous Percoll gradient (Sigma-Aldrich, Taufkirchen, Germany) was subsequently used to isolate polymorphonuclear leukocytes (PMNs). The cells were seeded at 5.0 × 106 cells/mL in RPMI medium (PAA, Traun, Austria) supplemented with 2 mM l-glutamin and 10% heat-inactivated, autologous donor serum. For infection, an Mtb H37Rv suspension was pelleted and washed twice in RPMI. Upon resuspension in RPMI, mycobacteria were opsonized with fresh autologous donor serum for 30 min at RT. PMNs were infected with an MOI of 3 and incubated for 6 h at 37 °C with 5% CO2. To assess the infection rate of the PMN culture, CFUs were determined as described in the Supporting Information. For targeted lipid analysis, the second aliquot of 1 mL cell culture was washed twice with PBS and finally harvested by centrifugation for 10 min at 15,000g at 4 °C. Cell pellets were resuspended in 50 μL of PBS and transferred into tubes containing 270 μL of methanol + 0.184% BHT (w/v) and incubated overnight at 4 °C to inactivate the bacteria. Lipid extraction was afterward continued according to the extraction protocol described below.
Mycobacterial Infection of Mice
Interleukin (IL)-13-overexpressing (IL-13TG) and wild-type littermates (IL-13WT) on a BALB/c genetic background20 were bred under specific-pathogen-free conditions. For infection experiments, 8 to 12 week old mice were transferred to the BSL3 facility at the Research Center Borstel and kept in individually ventilated cages. Mice were infected with a low dose of appr. 100 CFU of Mtb H37Rv via aerosol, and bacterial loads in lungs were determined at 42, 62, and 139 days after infection as described earlier.57 To enable standardized lipid extraction of lung tissues, the earlier described method27 was modified (for details, see the Supporting Information). Briefly, tissue homogenate aliquots containing 0.012 g wet weight were extracted, and the total protein content was determined after MTBE extraction. Normalized CFUs for each sample were calculated in concordance with the volume of each tissue homogenate aliquot used for lipid extraction (data file S2). Lipid extraction was performed according to a customized MTBE-based extraction as described earlier (ref (27), see details in the Supporting Information).
Description of Patient Cohort
Patients with pulmonary TB (non-M/XDR-TB and M/XDR-TB) were enrolled at five respiratory care centers in Germany as previously described.51,58 The study inclusion criterion was active pulmonary TB confirmed by a molecular test (GeneXpert, Cepheid, Sunnyvale, USA). Written informed consent was obtained from all study patients. Blood for PBMC analysis was sampled before therapy start (t0: defined as time point 0–14 days post diagnosis); when this was clinically not possible, the number of days after the ideal baseline sampling was recorded (data file S4, S5). Subsequently, samples were taken at predefined time points throughout drug treatment. Patient therapy outcomes were assessed using the TBnet simplified outcome criteria2 and WHO-defined criteria.59
Healthy Cohort of the Research Center Borstel
A total of 39 healthy adult Caucasian volunteers were enrolled at the Center for Clinical Studies, Research Center Borstel. Due to defined exclusion criteria, no subject was treated with systemic steroids during the last month and antibiotics in the previous 2 months, suffered from asthma, COPD (stage GOLD III/IV), airway infections during the previous month, any immunosuppression, diabetes mellitus, and previous or active TB, and was pregnant or breastfeeding.
Ethic Statements
Animal experiments were approved by the Ministry of Energy, Agriculture, Environment, Nature and Digitization of the state of Schleswig-Holstein, Germany (approval no. 69-6/16).
Study approval for human samples from active TB patients (AZ 12-233) and healthy volunteers (AZ 15-194) was granted by the Ethics Committee of the University of Lübeck.
For isolation of human neutrophils, written consent approving and authorizing the use of the volunteers’ material for research purposes was obtained from all donors. The Ethics Committee of the University of Lübeck approved isolation of and experimentation with human peripheral blood cells (AZ12-202A and AZ19-071).
Isolation of PBMC Samples
Blood for isolation of mononuclear cells (PBMC) was sampled in a CPT vacutainer (BD, Heidelberg, Germany). PBMCs were isolated according to the manufacturer’s instructions from heparinized whole blood. PBMCs were harvested from the interface, washed twice in PBS (Biochrom, Berlin, Germany) (250g, 15 min, 4 °C), and counted (Casy2, Schärfe System, Reutlingen, Germany). Subsequently, cell pellets were stored at −80 °C prior to MS analysis.
Targeted Quantitation of TSA-Containing PIs of PBMC
PBMC pellets containing either 1 × 106, 2 × 106, or 5 × 106 cells were washed with PBS and afterwards taken up in 50 μL of water. After thorough mixing of the samples, an internal standard mix was added (Supporting Information, Table S3). All samples were extracted following a customized MTBE protocol.27,60 Briefly, first, 270 μL of MeOH containing 3% acetic acid was added. Second, 1 mL of MTBE was added to the mixture, resulting in a one-phase system that was incubated at RT for 1 h under constant shaking. For separation of lipids from hydrophilic components, phase separation was induced by the addition of 250 μL of water. After an additional incubation period of 5 min, the mixture was centrifuged for 2 min at RT at 13,400g. The upper organic phase (800 μL) was transferred into a new tube. The samples were extracted a second time under the same conditions. The combined organic phase was dried in a vacuum concentrator and subsequently dissolved in 50 μL of storage solution (CHCl3/MeOH/H2O, 60/30/4.5, v/v/v). Lipid extracts were stored until analysis at −80 °C. Shotgun lipidomics was performed as reported earlier27 using the TriVersa NanoMate (Advion, Ithaca, USA) as the nano-electrospray source coupled to an Orbitrap Q Exactive Plus (Thermo Fisher Scientific, Bremen, Germany) mass spectrometer. A targeted analysis was performed over a time period of 5 min in the negative-ion mode for PIs listed in Supporting Information Table S2. Lipid identification was performed with LipidXplorer.61,62 Lipid annotation was performed according to the fatty acyl/alkyl level as described earlier (ref (63), see details in Supporting Information, Figure S4). Quantities of TSA-containing PIs were calculated in relation to the response of the FA 18:1 (d7) fragment ion of the internal standard PI 15:0–18:1 (d7) (828.5625 → 288.2925).
Acknowledgments
The authors acknowledge the excellent technical assistance of Birgit Kullmann, Romina Pritzkow, Franziska Daduna, Jessica Hofmeister, and the RCB blood donation service. We thank Andrea Glaewe, Lenka Krabbe, and Johanna Doehling for recruiting healthy volunteers and collecting samples. We acknowledge the excellent technical assistance of Doreen Beyer, Carolin Golin, Svenja Goldenbaum, and Alexandra Hölscher. We are also grateful to Anja Walter and Marion Schuldt for supplying and cleaning the lab and to Ilka Monath, Christine Keller, Sarah Vieten, and Gerhard Schultheiß for organizing the animal facilities in Borstel and Kiel. We thank Prof. Dr. S. Niemann for critical review and discussion on the manuscript. The BioMaterialBank Nord is supported by the German Center for Lung Research and member of popgen 2.0 network (P2N). Figure S12 and TOC were created using icons from BioRender.com.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsinfecdis.2c00075.
Experimental methods; supporting results; figures and tables; and MSn exemplary spectra (PDF)
Detection of TSA-containing lipids in MTBC (XLSX)
Determination of CFU and PI 16:0_19:0 in murine lung tissue (XLSX)
Analysis of PBMCs of 39 healthy controls (XLSX)
Analysis of PBMCs from TB patients at therapy start (XLSX)
Data for comparison of TB patients at t0 and tE (XLSX)
Author Present Address
§§ Thermo Fisher Scientific, Hanna-Kunath Str. 11, 28199 Bremen, Germany
Author Present Address
∥∥ Eurofins Professional Scientific Services Germany GmbH, Behringstr. 6/8 82152 Planegg, Germany.
Author Present Address
⊥⊥ Clinical Research Department, London School of Hygiene and Tropical Medicine, Keppel Street, London, WC1E 7HT, United Kingdom.
Author Present Address
## Karlsruhe Institute of Technology, 76344 Eggenstein-Leopoldshafen, Germany.
Author Contributions
△ First authors J.B., J.H., and F.M. (listed alphabetically) contributed equally; senior authors N.R. and D.S. contributed equally. J.H., N.R., and D.S. conceived and designed the study. N.Z., H.K., F.M., L.L., M.W., and V.S. performed lipidomics experiments and contributed to the data analysis. J.B. and L.L. performed cell culture experiments. F.M. and C. Hölscher performed mice experiments. N.R., S.H., and S.M. performed mycobacterial cultivation and selected strains of MTBC. K.K. designed the bacteriological test system for sputum analysis, and H.K. performed the lipid analysis. Recruitment and sample collection for TB patients were organized by J.H., B.K., and C. Herzmann. Recruitment and sample collection for healthy individual were managed by C. Herzmann and K.D. Collection and evaluation of medical data were overseen by P.S.C., B.K., C. Herzmann, K.G., M.R., and J.H.; J.B., J.H., F.M., N.G., N.R., and D.S. drafted the manuscript. All authors edited and commented on the final manuscript.
The authors are very grateful for funding of the German Center for Infection Research (DZIF) of the “thematic translational unit tuberculosis” (TTU TB; CHö: TTU 02.705; NR: TTU 02.806; 02.810; DS: TTU 02.704-1, 02.811). DS acknowledges the financial support of the German Network for Bioinformatics Infrastructure (de.NBI) for the project LIFS2 (FKZ 031L0108B).
The authors declare no competing financial interest.
Author Status
¶¶ Michael Weinkauf passed away unexpectedly in March, 2021. We lost a highly respected colleague who was also instrumental for the implementation of this study.
Notes
All data associated with this study are present in the paper or the Supporting Information.
Supplementary Material
References
- World Health Organization . Global Tuberculosis Report 2020; World Health Organization: Geneva, 2020.
- Günther G.; Lange C.; Alexandru S.; Altet N.; Avsar K.; Bang D.; Barbuta R.; Bothamley G.; Ciobanu A.; Crudu V.; Danilovits M.; Dedicoat M.; Duarte R.; Gualano G.; Kunst H.; de Lange W.; Leimane V.; Magis-Escurra C.; McLaughlin A.-M.; Muylle I.; Polcová V.; Popa C.; Rumetshofer R.; Skrahina A.; Solodovnikova V.; Spinu V.; Tiberi S.; Viiklepp P.; van Leth F.; for T. Treatment Outcomes in Multidrug-Resistant Tuberculosis. N. Engl. J. Med. 2016, 375, 1103–1105. 10.1056/nejmc1603274. [DOI] [PubMed] [Google Scholar]
- Schnippel K.; Ndjeka N.; Maartens G.; Meintjes G.; Master I.; Ismail N.; Hughes J.; Ferreira H.; Padanilam X.; Romero R.; Te Riele J.; Conradie F. Effect of bedaquiline on mortality in South African patients with drug-resistant tuberculosis: a retrospective cohort study. Lancet Respir. Med. 2018, 6, 699–706. 10.1016/s2213-2600(18)30235-2. [DOI] [PubMed] [Google Scholar]
- Heyckendorf J.; Olaru I. D.; Ruhwald M.; Lange C. Getting personal perspectives on individualized treatment duration in multidrug-resistant and extensively drug-resistant tuberculosis. Am. J. Respir. Crit. Care Med. 2014, 190, 374–383. 10.1164/rccm.201402-0363pp. [DOI] [PubMed] [Google Scholar]
- Chatterjee D. The mycobacterial cell wall: structure, biosynthesis and sites of drug action. Curr. Opin. Chem. Biol. 1997, 1, 579–588. 10.1016/s1367-5931(97)80055-5. [DOI] [PubMed] [Google Scholar]
- Jackson M. The mycobacterial cell envelope-lipids. Cold Spring Harbor Perspect. Med. 2014, 4, a021105. 10.1101/cshperspect.a021105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffé M.; Draper P. The envelope layers of mycobacteria with reference to their pathogenicity. Adv. Microb. Physiol. 1998, 39, 131–203. 10.1016/s0065-2911(08)60016-8. [DOI] [PubMed] [Google Scholar]
- Layre E.; Al-Mubarak R.; Belisle J. T.; Branch Moody D. Mycobacterial Lipidomics. Microbiol. Spectrum 2014, 2, 1–19. 10.1128/microbiolspec.MGM2-0033-2013. [DOI] [PubMed] [Google Scholar]
- Sartain M. J.; Dick D. L.; Rithner C. D.; Crick D. C.; Belisle J. T. Lipidomic analyses of Mycobacterium tuberculosis based on accurate mass measurements and the novel “Mtb LipidDB”. J. Lipid Res. 2011, 52, 861–872. 10.1194/jlr.m010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köfeler H. C.; Fauland A.; Rechberger G. N.; Trötzmüller M. Mass spectrometry based lipidomics: an overview of technological platforms. Metabolites 2012, 2, 19–38. 10.3390/metabo2010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triebl A.; Hartler J.; Trötzmüller M.; Köfeler H. C. Lipidomics: Prospects from a technological perspective. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2017, 1862, 740–746. 10.1016/j.bbalip.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shui G.; Bendt A. K.; Jappar I. A.; Lim H. M.; Laneelle M.; Hervé M.; Via L. E.; Chua G. H.; Bratschi M. W.; Zainul Rahim S. Z.; Michelle A. L. T.; Hwang S. H.; Lee J. S.; Eum S. Y.; Kwak H. K.; Daffé M.; Dartois V.; Michel G.; Barry C. E. 3rd; Wenk M. R. Mycolic acids as diagnostic markers for tuberculosis case detection in humans and drug efficacy in mice. EMBO Mol. Med. 2012, 4, 27–37. 10.1002/emmm.201100185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk R.; Kowalski K.; Janiszewska-Drobinska B.; Druszczyńska M. Rapid method for Mycobacterium tuberculosis identification using electrospray ionization tandem mass spectrometry analysis of mycolic acids. Diagn. Microbiol. Infect. Dis. 2013, 76, 298–305. 10.1016/j.diagmicrobio.2013.03.025. [DOI] [PubMed] [Google Scholar]
- Blanc L.; Lenaerts A.; Dartois V.; Prideaux B. Visualization of Mycobacterial Biomarkers and Tuberculosis Drugs in Infected Tissue by MALDI-MS Imaging. Anal. Chem. 2018, 90, 6275–6282. 10.1021/acs.analchem.8b00985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbek A.; Aktas O. Identification of three strains of Mycobacterium species isolated from clinical samples using fatty acid methyl ester profiling. J. Int. Med. Res. 2003, 31, 133–140. 10.1177/147323000303100210. [DOI] [PubMed] [Google Scholar]
- Brandenburg J.; Marwitz S.; Tazoll S. C.; Waldow F.; Kalsdorf B.; Vierbuchen T.; Scholzen T.; Gross A.; Goldenbaum S.; Hölscher A.; Hein M.; Linnemann L.; Reimann M.; Kispert A.; Leitges M.; Rupp J.; Lange C.; Niemann S.; Behrends J.; Goldmann T.; Heine H.; Schaible U. E.; Hölscher C.; Schwudke D.; Reiling N. WNT6-ACC2-induced accumulation of triacylglycerol rich lipid droplets is exploited by M. tuberculosis. J. Clin. Invest. 2021, 131, e141833 10.1172/jci141833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallenga T.; Repnik U.; Corleis B.; Eich J.; Reimer R.; Griffiths G. W.; Schaible U. E. M. tuberculosis-Induced Necrosis of Infected Neutrophils Promotes Bacterial Growth Following Phagocytosis by Macrophages. Cell Host Microbe 2017, 22, 519–530. 10.1016/j.chom.2017.09.003. [DOI] [PubMed] [Google Scholar]
- Kramnik I.; Beamer G. Mouse models of human TB pathology: roles in the analysis of necrosis and the development of host-directed therapies. Semin. Immunopathol. 2016, 38, 221–237. 10.1007/s00281-015-0538-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrey F.; Löwe D.; Kuhlmann S.; Kaiser P.; Moura-Alves P.; Krishnamoorthy G.; Lozza L.; Maertzdorf J.; Skrahina T.; Skrahina A.; Gengenbacher M.; Nouailles G.; Kaufmann S. H. E. Humanized Mouse Model Mimicking Pathology of Human Tuberculosis for in vivo Evaluation of Drug Regimens. Front. Immunol. 2019, 10, 89. 10.3389/fimmu.2019.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitmann L.; Abad Dar M.; Schreiber T.; Erdmann H.; Behrends J.; McKenzie A. N.; Brombacher F.; Ehlers S.; Hölscher C. The IL -13/ IL-4R α axis is involved in tuberculosis-associated pathology. J. Pathol. 2014, 234, 338–350. 10.1002/path.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölscher C.; Heitmann L.; Owusu-Dabo E.; Horstmann R. D.; Meyer C. G.; Ehlers S.; Thye T. A Mutation in IL4RA Is Associated with the Degree of Pathology in Human TB Patients. Mediat. Inflamm. 2016, 2016, 4245028. 10.1155/2016/4245028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends J.; Renauld J.-C.; Ehlers S.; Hölscher C. IL-22 Is Mainly Produced by IFNγ-Secreting Cells but Is Dispensable for Host Protection against Mycobacterium tuberculosis Infection. PLoS One 2013, 8, e57379 10.1371/journal.pone.0057379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begum H.; Li B.; Shui G.; Cazenave-Gassiot A.; Soong R.; Ong R. T.-H.; Little P.; Teo Y.-Y.; Wenk M. R. Discovering and validating between-subject variations in plasma lipids in healthy subjects. Sci. Rep. 2016, 6, 19139. 10.1038/srep19139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua E. C.-P.; Shui G.; Lee I. T.-G.; Lau P.; Tan L.-C.; Yeo S.-C.; Lam B. D.; Bulchand S.; Summers S. A.; Puvanendran K.; Rozen S. G.; Wenk M. R.; Gooley J. J. Extensive diversity in circadian regulation of plasma lipids and evidence for different circadian metabolic phenotypes in humans. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 14468–14473. 10.1073/pnas.1222647110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egen J. G.; Rothfuchs A. G.; Feng C. G.; Winter N.; Sher A.; Germain R. N. Macrophage and T cell dynamics during the development and disintegration of mycobacterial granulomas. Immunity 2008, 28, 271–284. 10.1016/j.immuni.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boom W. H.; Schaible U. E.; Achkar J. M. The knowns and unknowns of latent Mycobacterium tuberculosis infection. J. Clin. Investig. 2021, 131, e136222 10.1172/jci136222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers L. F.; Schwudke D. Shotgun Lipidomics Approach for Clinical Samples. Methods Mol. Biol. 2018, 1730, 163–174. 10.1007/978-1-4939-7592-1_12. [DOI] [PubMed] [Google Scholar]
- Machida S.; Bakku R. K.; Suzuki I. Expression of Genes for a Flavin Adenine Dinucleotide-Binding Oxidoreductase and a Methyltransferase from Mycobacterium chlorophenolicum Is Necessary for Biosynthesis of 10-Methyl Stearic Acid from Oleic Acid in Escherichia coli. Front. Microbiol. 2017, 8, 2061. 10.3389/fmicb.2017.02061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meena L. S.; Kolattukudy P. E. Expression and characterization of Rv0447c product, potentially the methyltransferase involved in tuberculostearic acid biosynthesis inMycobacterium tuberculosis. Biotechnol. Appl. Biochem. 2013, 60, 412–416. 10.1002/bab.1112. [DOI] [PubMed] [Google Scholar]
- Lawn S. D.; Edwards D. J.; Kranzer K.; Vogt M.; Bekker L.-G.; Wood R. Urine lipoarabinomannan assay for tuberculosis screening before antiretroviral therapy diagnostic yield and association with immune reconstitution disease. AIDS 2009, 23, 1875–1880. 10.1097/qad.0b013e32832e05c8. [DOI] [PubMed] [Google Scholar]
- Correia-Neves M.; Froberg G.; Korshun L.; Viegas S.; Vaz P.; Ramanlal N.; Bruchfeld J.; Hamasur B.; Brennan P.; Kallenius G. Biomarkers for tuberculosis: the case for lipoarabinomannan. ERJ Open Res. 2019, 5, 00115-2018. 10.1183/23120541.00115-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traunmüller F.; Alexander Zeitlinger M.; Stoiser B.; Lagler H.; Abdulla Abdel Salam H.; Presterl E.; Graninger W. Circulating tuberculostearic acid in tuberculosis patients. Scand. J. Infect. Dis. 2003, 35, 790–793. 10.1080/00365540310017032. [DOI] [PubMed] [Google Scholar]
- Odham G.; Larsson L.; Mårdh P. A. Demonstration of tuberculostearic acid in sputum from patients with pulmonary tuberculosis by selected ion monitoring. J. Clin. Invest. 1979, 63, 813–819. 10.1172/jci109380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourão M. P. B.; Kuijper S.; Dang N. A.; Walters E.; Janssen H.-G.; Kolk A. H. J. Direct detection of Mycobacterium tuberculosis in sputum: A validation study using solid phase extraction-gas chromatography-mass spectrometry. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2016, 1012–1013, 50–54. 10.1016/j.jchromb.2015.12.023. [DOI] [PubMed] [Google Scholar]
- Muranishi H.; Nakashima M.; Isobe R.; Ando T.; Shigematsu N. Measurement of tuberculostearic acid in sputa, pleural effusions, and bronchial washings. A clinical evaluation for diagnosis of pulmonary tuberculosis. Diagn. Microbiol. Infect. Dis. 1990, 13, 235–240. 10.1016/0732-8893(90)90065-4. [DOI] [PubMed] [Google Scholar]
- Muranishi H.; Nakashima M.; Hirano H.; Saitoh T.; Takahashi H.; Tanaka K.; Miyazaki M.; Yagawa K.; Shigematsu N. Simultaneous measurements of adenosine deaminase activity and tuberculostearic acid in pleural effusions for the diagnosis of tuberculous pleuritis. Intern. Med. 1992, 31, 752–755. 10.2169/internalmedicine.31.752. [DOI] [PubMed] [Google Scholar]
- Yorgancioglu A.; Akin M.; Dereli S.; Aktogu S.; Ilis Z.; Sezgin A. The diagnostic value of tuberculostearic acid in tuberculous pleural effusions. Monaldi Arch. Chest Dis. 1996, 51, 108–111. [PubMed] [Google Scholar]
- Takayama K.; Wang C.; Besra G. S. Pathway to synthesis and processing of mycolic acids in Mycobacterium tuberculosis. Clin. Microbiol. Rev. 2005, 18, 81–101. 10.1128/cmr.18.1.81-101.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nataraj V.; Varela C.; Javid A.; Singh A.; Besra G. S.; Bhatt A. Mycolic acids: deciphering and targeting the Achilles’ heel of the tubercle bacillus. Mol. Microbiol. 2015, 98, 7–16. 10.1111/mmi.13101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spargo B. J.; Crowe L. M.; Ioneda T.; Beaman B. L.; Crowe J. H. Cord factor (alpha,alpha-trehalose 6,6’-dimycolate) inhibits fusion between phospholipid vesicles. Proc. Natl. Acad. Sci. U.S.A. 1991, 88, 737–740. 10.1073/pnas.88.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indrigo J.; Hunter R. L. Jr.; Actor J. K. Cord factor trehalose 6,6′-dimycolate (TDM) mediates trafficking events during mycobacterial infection of murine macrophages. Microbiology 2003, 149, 2049–2059. 10.1099/mic.0.26226-0. [DOI] [PubMed] [Google Scholar]
- Axelrod S.; Oschkinat H.; Enders J.; Schlegel B.; Brinkmann V.; Kaufmann S. H. E.; Haas A.; Schaible U. E. Delay of phagosome maturation by a mycobacterial lipid is reversed by nitric oxide. Cell. Microbiol. 2008, 10, 1530–1545. 10.1111/j.1462-5822.2008.01147.x. [DOI] [PubMed] [Google Scholar]
- Fischer K.; Chatterjee D.; Torrelles J.; Brennan P. J.; Kaufmann S. H. E.; Schaible U. E. Mycobacterial lysocardiolipin is exported from phagosomes upon cleavage of cardiolipin by a macrophage-derived lysosomal phospholipase A2. J. Immunol. 2001, 167, 2187–2192. 10.4049/jimmunol.167.4.2187. [DOI] [PubMed] [Google Scholar]
- Barkan D.; Hedhli D.; Yan H.-G.; Huygen K.; Glickman M. S. Mycobacterium tuberculosis lacking all mycolic acid cyclopropanation is viable but highly attenuated and hyperinflammatory in mice. Infect. Immun. 2012, 80, 1958–1968. 10.1128/iai.00021-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang N. A.; Kuijper S.; Walters E.; Claassens M.; van Soolingen D.; Vivo-Truyols G.; Janssen H.-G.; Kolk A. H. J. Validation of Biomarkers for Distinguishing Mycobacterium tuberculosis from Non-Tuberculous Mycobacteria Using Gas Chromatography–Mass Spectrometry and Chemometrics. PLoS One 2013, 8, e76263 10.1371/journal.pone.0076263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilleron M.; Quesniaux V. F. J.; Puzo G. Acylation State of the Phosphatidylinositol Hexamannosides from Mycobacterium bovis Bacillus Calmette Guérin and Mycobacterium tuberculosis H37Rv and Its Implication in Toll-like Receptor Response. J. Biol. Chem. 2003, 278, 29880–29889. 10.1074/jbc.m303446200. [DOI] [PubMed] [Google Scholar]
- Holst O.; P Moran A.; Brennan P. J.. Overview of the glycosylated components of the bacterial cell envelope. In Microbial Glycobiology; Holst O., Brennan P. J., von Itzstein M. M., Eds.; Academic Press, 2010; pp 3–13. [Google Scholar]
- Angala S. K.; Belardinelli J. M.; Huc-Claustre E.; Wheat W. H.; Jackson M. The cell envelope glycoconjugates ofMycobacterium tuberculosis. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 361–399. 10.3109/10409238.2014.925420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich S. O.; Rachow A.; Saathoff E.; Singh K.; Mangu C. D.; Dawson R.; Phillips P. P.; Venter A.; Bateson A.; Boehme C. C.; Heinrich N.; Hunt R. D.; Boeree M. J.; Zumla A.; McHugh T. D.; Gillespie S. H.; Diacon A. H.; Hoelscher M.; Pan A. Assessment of the sensitivity and specificity of Xpert MTB/RIF assay as an early sputum biomarker of response to tuberculosis treatment. Lancet Respir. Med. 2013, 1, 462–470. 10.1016/s2213-2600(13)70119-x. [DOI] [PubMed] [Google Scholar]
- Dorman S. E.; Schumacher S. G.; Alland D.; Nabeta P.; Armstrong D. T.; King B.; Hall S. L.; Chakravorty S.; Cirillo D. M.; Tukvadze N.; Bablishvili N.; Stevens W.; Scott L.; Rodrigues C.; Kazi M. I.; Joloba M.; Nakiyingi L.; Nicol M. P.; Ghebrekristos Y.; Anyango I.; Murithi W.; Dietze R.; Lyrio Peres R.; Skrahina A.; Auchynka V.; Chopra K. K.; Hanif M.; Liu X.; Yuan X.; Boehme C. C.; Ellner J. J.; Denkinger C. M.; Dorman S. E.; Schumacher S. G.; Alland D.; Nabeta P.; Armstrong D. T.; King B.; Hall S. L.; Chakravorty S.; Cirillo D. M.; Tukvadze N.; Bablishvili N.; Stevens W.; Scott L.; Rodrigues C.; Kazi M. I.; Joloba M.; Nakiyingi L.; Nicol M. P.; Ghebrekristos Y.; Anyango I.; Murithi W.; Dietze R.; Peres R. L.; Skrahina A.; Auchynka V.; Chopra K. K.; Hanif M.; Liu X.; Yuan X.; Boehme C. C.; Ellner J. J.; Denkinger C. M.; Manabe Y. C.; Hom D.; Aspindzelashvili R.; David A.; Surve U.; Kamulegeya L. H.; Nabweyambo S.; Surtie S.; Hapeela N.; Cain K. P.; Agaya J.; McCarthy K. D.; Marques-Rodrigues P.; Schmidt Castellani L. G.; Almeida P. S.; de Aguiar P. P. L.; Solodovnikova V.; Ruan X.; Liang L.; Zhang G.; Zhu H.; Xie Y. Xpert MTB/RIF Ultra for detection of Mycobacterium tuberculosis and rifampicin resistance: a prospective multicentre diagnostic accuracy study. Lancet Infect. Dis. 2018, 18, 76–84. 10.1016/s1473-3099(17)30691-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyckendorf J.; van Leth F.; Avsar K.; Glattki G.; Günther G.; Kalsdorf B.; Müller M.; Olaru I. D.; Rolling T.; Salzer H. J. F.; Schuhmann M.; Terhalle E.; Lange C. Treatment responses in multidrug-resistant tuberculosis in Germany. IJTLD 2018, 22, 399–406. 10.5588/ijtld.17.0741. [DOI] [PubMed] [Google Scholar]
- Yamada H.; Mitarai S.; Aguiman L.; Matsumoto H.; Fujiki A. Preparation of mycobacteria-containing artificial sputum for TB panel testing and microscopy of sputum smears. IJTLD 2006, 10, 899–905. [PubMed] [Google Scholar]
- Michelucci A.; Cordes T.; Ghelfi J.; Pailot A.; Reiling N.; Goldmann O.; Binz T.; Wegner A.; Tallam A.; Rausell A.; Buttini M.; Linster C. L.; Medina E.; Balling R.; Hiller K. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 7820–7825. 10.1073/pnas.1218599110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Goncalves R.; Mosser D. M.. The isolation and characterization of murine macrophages. Current Protocols in Immunology; Wiley, 2008; Chapter 14, Unit 14 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauser C.; Dallenga T.; Tchikov V.; Schaible U. E.; Schutze S.; Reiling N.. Immunomagnetic isolation of pathogen-containing phagosomes and apoptotic blebs from primary phagocytes. Current Protocols in Immunology; Wiley, 2014; Vol 105, pp 14 36 11–14 36 26. [DOI] [PubMed] [Google Scholar]
- Schaale K.; Brandenburg J.; Kispert A.; Leitges M.; Ehlers S.; Reiling N. Wnt6 Is Expressed in Granulomatous Lesions ofMycobacterium tuberculosis-Infected Mice and Is Involved in Macrophage Differentiation and Proliferation. J. Immunol. 2013, 191, 5182–5195. 10.4049/jimmunol.1201819. [DOI] [PubMed] [Google Scholar]
- Hölscher C.; Reiling N.; Schaible U. E.; Hölscher A.; Bathmann C.; Korbel D.; Lenz I.; Sonntag T.; Kröger S.; Akira S.; Mossmann H.; Kirschning C. J.; Wagner H.; Freudenberg M.; Ehlers S. Containment of aerogenicMycobacterium tuberculosis infection in mice does not require MyD88 adaptor function for TLR2, -4 and -9. Eur. J. Immunol. 2008, 38, 680–694. 10.1002/eji.200736458. [DOI] [PubMed] [Google Scholar]
- Heyckendorf J.; van Leth F.; Kalsdorf B.; Olaru I. D.; Gunther G.; Salzer H. J. F.; Terhalle E.; Rolling T.; Glattki G.; Muller M.; Schuhmann M.; Avsar K.; Lange C. Relapse-free cure from multidrug-resistant tuberculosis in Germany. Eur. Respir. J. 2018, 51, 1702122. 10.1183/13993003.02122-2017. [DOI] [PubMed] [Google Scholar]
- WHO . Definitions and Reporting Framework for Tuberculosis—2013; World Health Organization: Geneva, 2014.
- Matyash V.; Liebisch G.; Kurzchalia T. V.; Shevchenko A.; Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 2008, 49, 1137–1146. 10.1194/jlr.d700041-jlr200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog R.; Schwudke D.; Schuhmann K.; Sampaio J. L.; Bornstein S. R.; Schroeder M.; Shevchenko A. A novel informatics concept for high-throughput shotgun lipidomics based on the molecular fragmentation query language. Genome Biol. 2011, 12, R8. 10.1186/gb-2011-12-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog R.; Schuhmann K.; Schwudke D.; Sampaio J. L.; Bornstein S. R.; Schroeder M.; Shevchenko A. LipidXplorer: a software for consensual cross-platform lipidomics. PLoS One 2012, 7, e29851 10.1371/journal.pone.0029851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebisch G.; Vizcaíno J. A.; Köfeler H.; Trötzmüller M.; Griffiths W. J.; Schmitz G.; Spener F.; Wakelam M. J. O. Shorthand notation for lipid structures derived from mass spectrometry. J. Lipid Res. 2013, 54, 1523–1530. 10.1194/jlr.m033506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiling N.; Homolka S.; Kohl T. A.; Steinhäuser C.; Kolbe K.; Schütze S.; Brandenburg J. Shaping the niche in macrophages: Genetic diversity of the M. tuberculosis complex and its consequences for the infected host. Int. J. Med. Microbiol. 2018, 308, 118–128. 10.1016/j.ijmm.2017.09.009. [DOI] [PubMed] [Google Scholar]
- Homolka S.; Niemann S.; Russell D. G.; Rohde K. H. Functional genetic diversity among Mycobacterium tuberculosis complex clinical isolates: delineation of conserved core and lineage-specific transcriptomes during intracellular survival. PLoS Pathog. 2010, 6, e1000988 10.1371/journal.ppat.1000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerriegel S.; Koser C. U.; Niemann S. Phylogenetic polymorphisms in antibiotic resistance genes of the Mycobacterium tuberculosis complex. J. Antimicrob. Chemother. 2014, 69, 1205–1210. 10.1093/jac/dkt535. [DOI] [PubMed] [Google Scholar]
- Zelmer A.; Carroll P.; Andreu N.; Hagens K.; Mahlo J.; Redinger N.; Robertson B. D.; Wiles S.; Ward T. H.; Parish T.; Ripoll J.; Bancroft G. J.; Schaible U. E. A new in vivo model to test anti-tuberculosis drugs using fluorescence imaging. J. Antimicrob. Chemother. 2012, 67, 1948–1960. 10.1093/jac/dks161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll F.; Phelan J.; Hill-Cawthorne G. A.; Nair M. B.; Mallard K.; Ali S.; Abdallah A. M.; Alghamdi S.; Alsomali M.; Ahmed A. O.; Portelli S.; Oppong Y.; Alves A.; Bessa T. B.; Campino S.; Caws M.; Chatterjee A.; Crampin A. C.; Dheda K.; Furnham N.; Glynn J. R.; Grandjean L.; Minh Ha D.; Hasan R.; Hasan Z.; Hibberd M. L.; Joloba M.; Jones-López E. C.; Matsumoto T.; Miranda A.; Moore D. J.; Mocillo N.; Panaiotov S.; Parkhill J.; Penha C.; Perdigão J.; Portugal I.; Rchiad Z.; Robledo J.; Sheen P.; Shesha N. T.; Sirgel F. A.; Sola C.; Oliveira Sousa E.; Streicher E. M.; Helden P. V.; Viveiros M.; Warren R. M.; McNerney R.; Pain A.; Clark T. G. Genome-wide analysis of multi- and extensively drug-resistant Mycobacterium tuberculosis. Nat. Genet. 2018, 50, 307–316. 10.1038/s41588-017-0029-0. [DOI] [PubMed] [Google Scholar]
- Homolka S.; Projahn M.; Feuerriegel S.; Ubben T.; Diel R.; Nübel U.; Niemann S. High resolution discrimination of clinical Mycobacterium tuberculosis complex strains based on single nucleotide polymorphisms. PLoS One 2012, 7, e39855 10.1371/journal.pone.0039855. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.