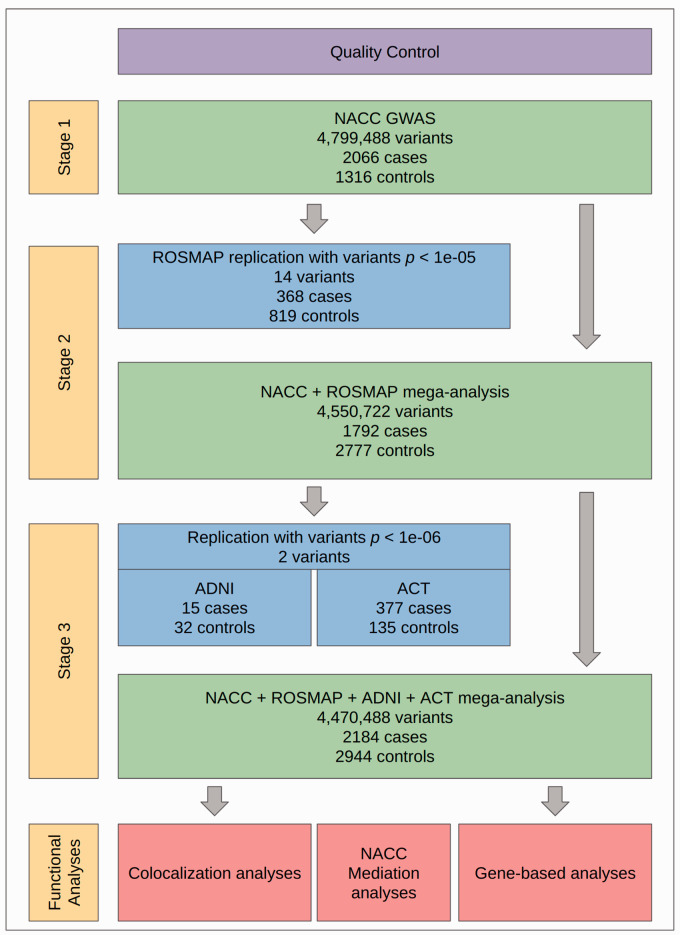
Figure 1.
Overall study design. First, quality control (QC) was performed on all neuropathological and genotype data sets used (see Methods). GWAS were performed across three stages. In Stage 1, GWAS was performed on NACC participants (n = 3382). In Stage 2, all variants with p < (k = 14) were analyzed separately in ROSMAP (n = 1187), and then NACC and ROSMAP participant data were merged for mega-analysis (n = 4569). In Stage 3, top variants from Stage 2 mega-analysis with p < (k = 2) were analyzed in two replication cohorts, ADNI (n = 47) and ACT (n = 512), and then NACC, ROSMAP, ADNI, and ACT data were merged for mega-analysis. Downstream functional analyses, consisting of colocalization and gene-based analyses, were then performed on Stage 3 mega-analysis results. Mediation analyses for HTN and DM were also performed using a subset of NACC participants with clinical data available (n = 1726).