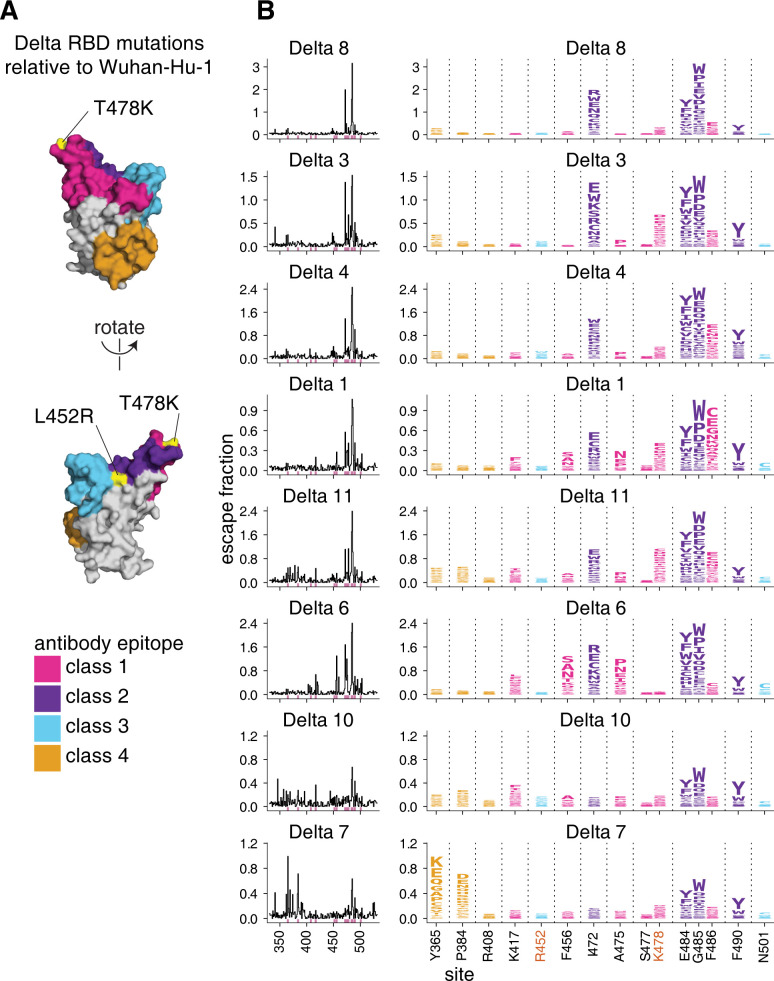
Fig 3. Complete antibody-escape maps for plasmas from individuals convalescent of primary Delta infections.
(A) The surface of the Wuhan-Hu-1 RBD is colored according to antibody epitope per the Barnes classification scheme [37], with the L452R and T478K mutations in the Delta variant highlighted in yellow (and labeled with red text in panel B). We define L452R to be in the class 3 epitope, and T478K in the class 1 epitope. (B) Complete maps of mutations that reduce binding of plasma antibodies to the Delta RBD from individuals convalescent of primary Delta infection. Sites of strong antibody escape (see Methods) for any of the 8 plasmas are highlighted with pink in the line plots at left and shown in the logo plots at right. Sites 452, 477, and 501 are included despite not being sites of strong escape due to their high frequency in circulating viral isolates. Site 408 is included to facilitate comparison to Delta breakthrough infection-elicited plasmas (Fig 5). Interactive versions of logo plots and structural visualizations are at https://jbloomlab.github.io/SARS-CoV-2-RBD_Delta/. Correlations between independent library replicates are in S5 Fig, and all escape scores are in S3 Data and online at https://github.com/jbloomlab/SARS-CoV-2-RBD_Delta/blob/main/results/supp_data/aggregate_raw_data.csv.