Figure 1.
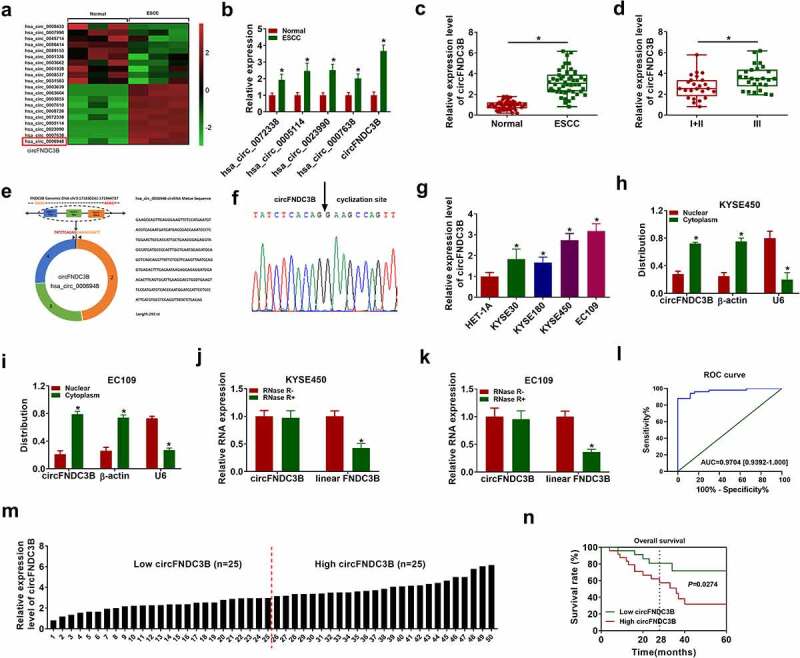
CircFNDC3B was overexpressed in ESCC. (a) The heat map of GSE131969 showed the expression of 20 circRNAs in normal and ESCC tissues. (b) Hsa_circ_0072338, hsa_circ_0005114, hsa_circ_0023990, hsa_circ_0007638, and circFNDC3B expression levels in normal and ESCC tissues were detected by RT-qPCR (two-way ANOVA). (c) CircFNDC3B expression in ESCC tissues (n = 39) and normal tissues (n = 39) was determined (Student’s t-test). (d) The expression of circFNDC3B was determined in patients with clinical stages I + II and III (Student’s t-test). (e,f) The information of circFNDC3B was presented, and Sanger sequencing confirmed the head-to-tail splicing junction of circFNDC3B. (g) The expression of circFNDC3B in ESCC cells (KYSE30, KYSE180, KYSE450, and EC109) and HET-1A cells was measured via RT-qPCR (one-way ANOVA). (h,i) The subcellular location of circFNDC3B in KYSE450 and EC109 cells was determined by RT-qPCR (two-way ANOVA). (j,k) CircFNDC3B and linear FNDC3B expression levels were determined after treatment of RNase R by RT-qPCR in KYSE450 and EC109 cells (two-way ANOVA). (l) The diagnostic effect was evaluated by ROC curve in ESCC patients (Clopper-Pearson). (m) ESCC patients were divided into low (n = 19) and high (n = 20) circFNDC3B expression groups according to the median value of circFNDC3B expression. (n) The survival rate was analyzed between low and high circFNDC3B expression groups in ESCC patients (Log-rank (Mantel-Cox) test). *P< 0.05.
