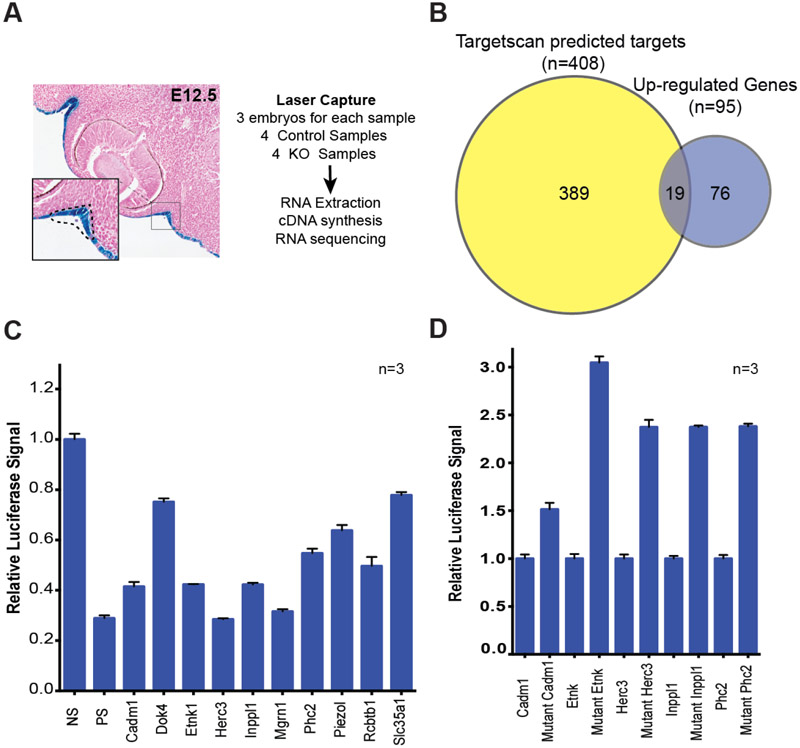
Fig. 5. RNA-seq of miR-205−/− tissues uncovers relevant miR-205 targets and pathways.
(A) Schematic overview of experimental method for tissue isolation and RNA sequencing. (B) Overlap between up-regulated genes and predicted miR-205 targets expressed in the surface ectoderm. (C) Luciferase UTR assays for subset of genes up-regulated in miR-205 knockout samples (SEM error bars). No binding site (NS), a perfect binding site (PS) control. (D) Luciferase UTR assays for miR-205 targets with wildtype UTRs (normalized to one) and mutated UTRs.