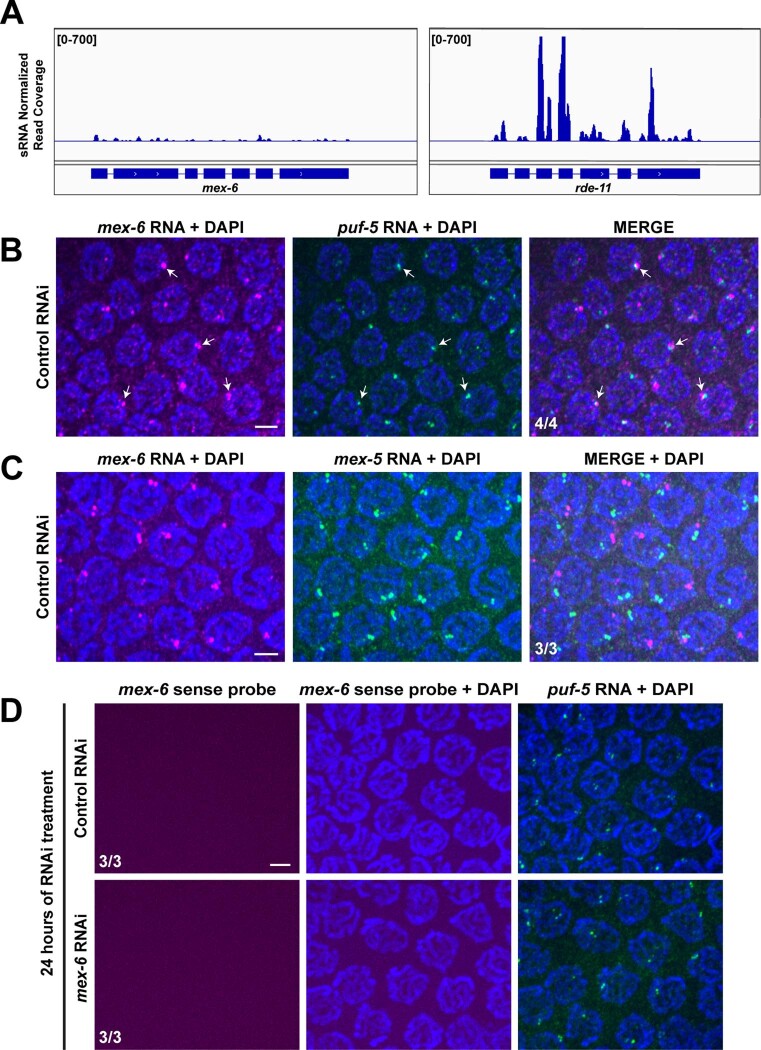
Extended Data Fig. 1. Characterization of the mex-6 transcript.
a) IGV genome browser views of sRNAseq reads (wild-type adult hermaphrodites) mapping to the mex-6 locus. rde-11 is a locus that is highly targeted by endogenous sRNAs52, shown here for comparison. Genome views are representative of two independent sequencing libraries. b) Maximum projection photomicrographs of pachytene nuclei showing DNA (blue, stained with DAPI), mex-6 RNA (magenta), and puf-5 RNA (green). The mex-6 and puf-5 loci are linked on Chromosome II and, as expected, exhibit closely linked nuclear puncta (arrows). Scale bar is 2.5 µm. Images are representative of 3 worms examined. Results were consistent across four independent FISH experiments. c) Same as in B but showing mex-6 (magenta) and mex-5 (green) RNAs. mex-6 and mex-5 are homologous loci on different chromosomes. Scale bar is 2.5 µm. Images are representative of 3 worms examined. Results were consistent across two independent FISH experiments. The mex-6 and mex-5 puncta do not co-localize, as expected, confirming the specificity of the FISH probes. d) Same as in B and C but using a mex-6 sense probe (magenta) and a puf-5 antisense probe (green). Scale bar is 2.5 µm. Images are representative of 3 worms examined. Results were consistent across two independent FISH experiments. As expected, the mex-6 sense probe does not detect any signal, confirming that the signals detected by the mex-6 antisense probe in panels B and C correspond to RNA and not DNA. Note also the lack of signal under RNAi conditions, suggesting that our FISH protocol does not detect sRNAs.