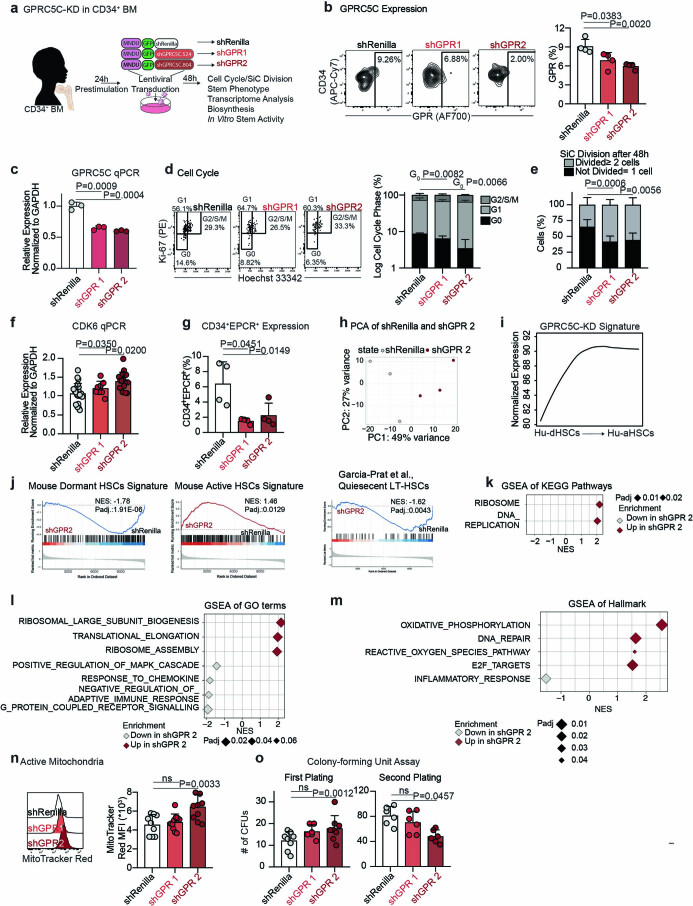
Extended Data Fig. 4.
a. Experimental design to knockdown and assess the impact of decreased GPRC5C levels in CD34+ BM. b. FACS validation of GPRC5C protein levels quantified in GFP+ BM cells. n = 4 experiments. c. qPCR of GPRC5C expression in GFP+ BM cells. Normalized to housekeeping gene GAPDH and shRenilla. n = 3 experiments. d. Cell cycle FACS analysis on GFP+ BM cells. Cell cycle phase is represented on log10 scale. n = 4 experiments. e. SiC division assay of GFP+ BM cells quantified after 48 hours in vitro culture. n = 4 experiments. f. qPCR of CDK6 expression in GFP+ BM cells. Normalized to housekeeping gene GAPDH and shRenilla. n = 8 experiments. g. Surface expression of CD34+EPCR+ in GFP+ BM cells. n = 4 experiments. h. PCA of RNA-seq data from shGPR 2 and shRenilla control. Based on top 5,000 variable genes. log2FC threshold ≥ 0.5, padj. < 0.1. i. Smoothed pseudo-temporal expression profile of the GPR-KD signature in LT-HSCs. Smoothed profiles were computed by local regression. The y-axis of the smoothed pseudo-temporal expression profiles indicates normalized expression. j. GSEA of Mouse Dormant HSCs, Mouse Active HSCs, and Quiescent LT-HSCs from García-Prat et al., signature in shGPR 2 and shRenilla control. k. GSEA of KEGG pathways in shGPR 2 compared to shRenilla control. l. GSEA of GO terms in shGPR 2 compared to shRenilla control. m. GSEA of Hallmarks terms in shGPR 2 compared to shRenilla control. n. FACS measurement of MitoTracker Red MFI in GFP+ BM. n = 10 experiments. o. CFU assay of GFP+ cells from GPRC5C-KD and shRenilla control. Primary plating n = 8 experiments, except n = 6 experiments for shGPR1. Secondary plating n = 6 experiments. All data presented as mean ± SD. Statistical significance was determined using two-tailed t-test, except d and e (two-way ANOVA). GSEA performed with BH-adjusted p-values after adaptive multilevel splitting Monte Carlo approach. ns, not significant. n indicates the number of biological replicates. For all experiments, at least two independent experiments were performed. At least two human BM donors were used. Source numerical data are available in source data.