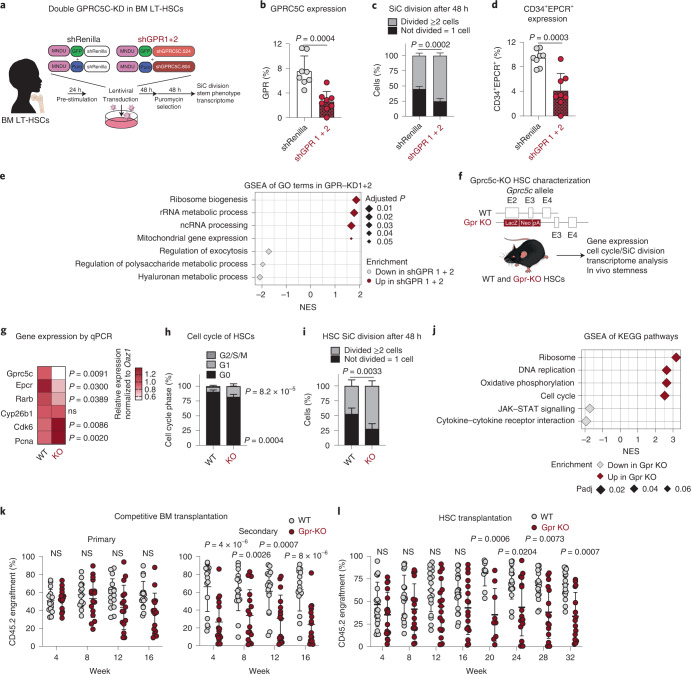
Fig. 3. Loss of GPRC5C impairs stemness and quiescence.
a, Experimental design to knock down and assess the impact of decreased GPRC5C levels in BM LT-HSCs. b, FACS validation of GPRC5C protein levels. n = 8 experiments. c, SiC division assay of GFP+ BM cells quantified after 48 h in vitro culture. n = 4 experiments for shGPR1 + 2 and n = 5 experiments for shRenilla. d, Surface expression of CD34+EPCR+ in GFP+ BM cells. n = 8 experiments. e, GSEA of GO terms in shGPR 1 + 2 compared with shRenilla control. f, Experimental design to assess functional impact of Gprc5c KO in mouse HSCs. Deletion of Gprc5c was achieved by the insertion of LacZ downstream of the translation initiation site in exon 2 of both alleles. g, Differential gene expression. Heat map representing median RNA expression from qPCR data (normalized to housekeeping gene Oaz1 and WT control). n = 8 biological replicates. h, FACS-based HSC cell cycle analysis with Ki-67 and DAPI. n = 5 biological replicates. i, SiC division assay of Gprc5c-KO and WT control HSCs quantified after 48 h in vitro culture. n = 4 biological replicates for Gpr KO and n = 5 biological replicates for WT. j, GSEA of KEGG pathways in Gprc5c-KO compared with WT control HSCs. k, PB analysis of competitive WBM chimaeras. CD45.2 cell percentage outcome in PB is presented. n = 16 biological replicates. l, PB analysis of HSC transplants. CD45.2 cell percentage outcome in PB is presented. n = 17 biological replicates for Gpr KO and n = 18 biological replicates for WT. All data presented as mean ± s.d. Statistical significance was determined by two-tailed t-test (b, d and g) or two-way ANOVA (c, h, I, k and l). n indicates number of replicates. GSEA was performed with BH-adjusted P values after adaptive multi-level splitting Monte Carlo approach. For all experiments, at least two independent experiments were performed. At least two human BM donors were used (b–d). Source numerical data are available in source data.