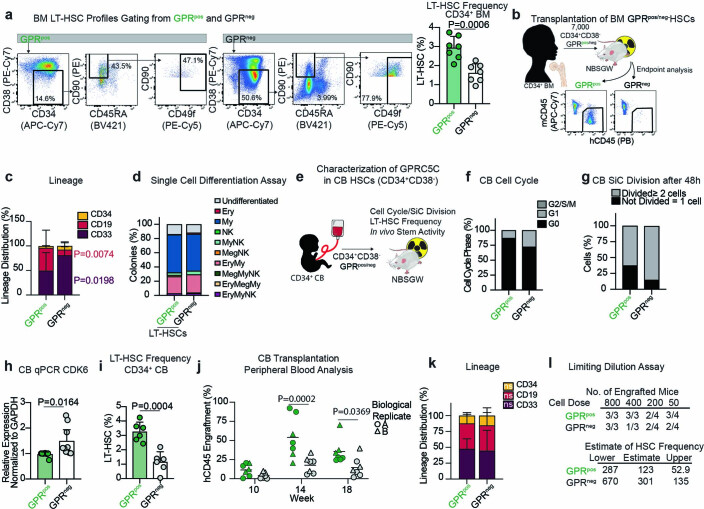
Extended Data Fig. 3.
a. Representative FACS gating scheme of LT-HSCs within GPRC5Cpos/neg population from CD34+ BM donors. Frequency of LT-HSCs within GPRC5Cpos/neg cells from CD34+ BM donors. n = 7 biological replicates. b. Experimental design to assess the in vivo reconstitution ability of BM GPRC5Cpos/neg-HSCs. c. Lineage distribution of GPRC5Cpos/neg-HSCs. Data presented refer to the distribution of myeloid (CD33), B-lymphoid (CD19) and immature surface phenotype (CD34). n = 9 biological replicates. d. Percentage of determined colony types derived from single BM GPRC5Cpos/neg -LT-HSCs following culture under differentiating conditions for three weeks. n = 2 biological replicates. e. Experimental design to characterize GPRC5C in human CB. f. FACS analysis of cell cycle from CB GPRC5Cpos/neg-HSCs. n = 2 biological replicates. g. SiC division assay of GPRC5Cpos/neg-HSCs from CB donors quantified after 48 hours in vitro culture. n = 2 biological replicates. h. qPCR of CDK6 expression from CB GPRC5Cpos/neg-HSCs. Normalized to housekeeping gene GAPDH and GPRC5Cpos-HSCs. n = 9 biological replicates. i. Frequency of LT-HSCs within GPRC5Cpos/neg cells from CD34+ CB donors. n = 6 biological replicates. j. Assessment of reconstitution dynamics. Engraftment was assessed by percentage of human CD45 cells in the PB. n = 6 biological replicates. k. Lineage contribution of CB GPRC5Cpos/neg-HSCs. Data presented refer to the distribution of myeloid (CD33), B-lymphoid (CD19), and immature surface phenotype (CD34). n = 6 biological replicates. l. Table showing the number of cells injected per dose, number of mice per dose, and the number of engrafted mice per dose. These data were used to calculate the HSC frequency seen in Fig. 2i. All data presented as mean ± SD. Statistical significance was determined using two-tailed t-test (a, h, i, j) or two-way ANOVA (c and k) and at least two independent experiments were performed. ns, not significant. n indicates number of replicates. Source numerical data are available in source data.