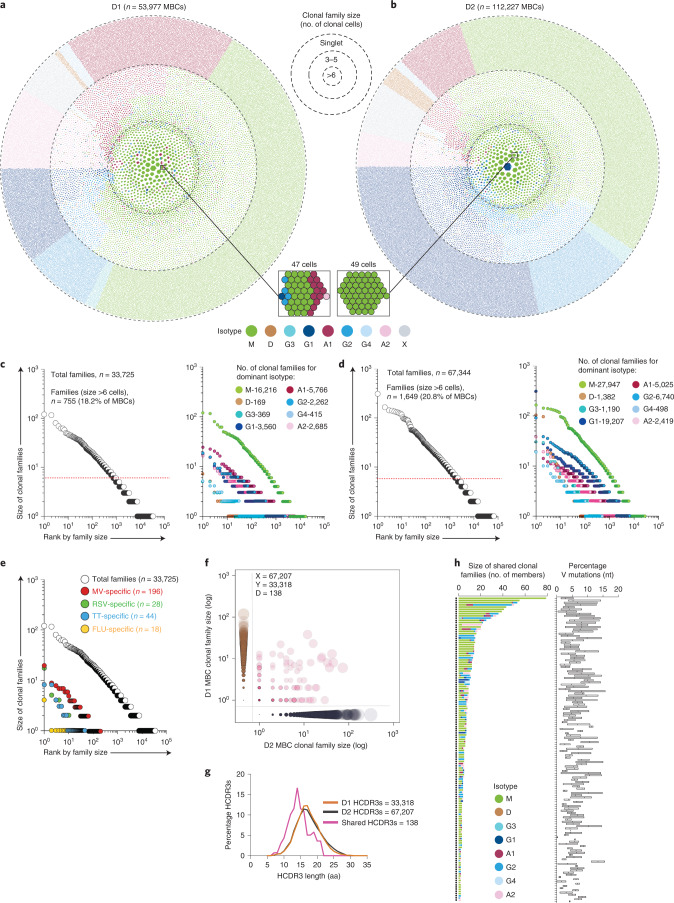
Fig. 1. Clonal structure of the memory B cell pool and convergent antibodies.
a,b, Honeycomb plots compile all memory B cells (MBCs) from D1 and D2 samples analyzed using a donor-specific database into clonal families; each cell is color-coded according to the isotype expressed and families are ranked according to size from center to periphery. Two representative clonal families comprising multiple or unique isotypes are highlighted. c,d, Waterfall plots represent the size distribution of all clonal families (left) and of families grouped according to the dominant (≥80% of cells) isotype (right) in D1 (c) and D2 (d). Dotted red lines separate families with more than six and six or fewer cells. Isotypes are color coded (x indicates isotype not determined). e, Size distribution of MBC families from D1 specific for the recall antigens measles virus (MV), respiratory syncytial virus (RSV), tetanus toxoid (TT) and influenza virus (FLU) determined by searching 328 antigen-specific antibody sequences (belonging to 286 clonal families) in the 10X Genomics database from D1. Size distribution of total MBC clonal families is included for comparison. f, Scatter-plot shows convergent clonotypes determined by jointly analyzing MBC sequences from D1 and D2 using publicly available reference sequences from the IMGT database, clonotypes shared between D1 and D2 (diagonal) and non-shared clonotypes (x and y axes) are ranked according to clonal family size. g, HCDR3 length distribution in shared and non-shared MBC clonotypes. h, Isotype usage and percent VH mutations in the shared clonotypes.