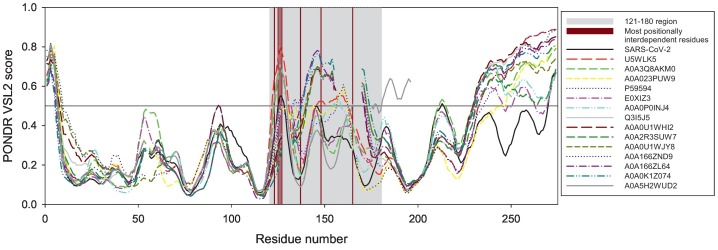
Fig. 4.
Per-residue intrinsic disorder predisposition of the 15 most evolutionary similar Spike RBDs evaluated by PONDR® VSL2 (33). Breaks in the disorder profiles correspond to the breaks in sequence alignments. In this analysis, disorder scores between 0.2 and 0.5 indicate flexible regions, whereas regions with the disorder scores ≥0.5 are expected to be intrinsically disordered. Position of the 121–180 region corresponding to the area with the highest co-evolutionary signal and thus with the maximal positionally interdependent residues is shown by light grey shading. Dark red bars show position of residues, which are most positionally interdependent (123, 125, 126, 127, 137, 148, and 165). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)