FIGURE 4.
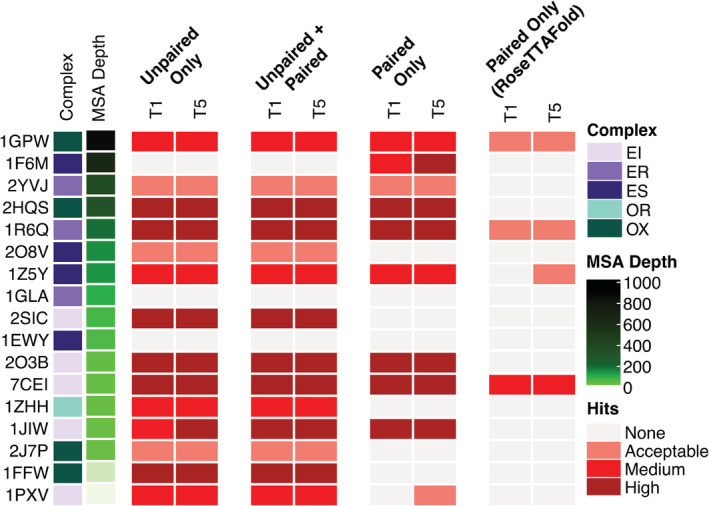
The impact of MSA pairing on prediction accuracy. MSAs were generated using MMseqs2 using the “advanced” interface of ColabFold. 29 Pairing was performed in ColabFold on a total of 17 cases whose ligand and receptor proteins come from the same prokaryotic organism. Cases in the heatmap were sorted by the paired MSA depth (N eff; see Material and Methods for details) from the largest to the smallest values. Structural predictions were generated with the “advanced” interface of ColabFold, and RoseTTAFold 28 (through the Robetta server). All models were assessed for near‐native predictions within the top‐ranked (T1) and top 5 (T5) models using CAPRI criteria. Complex category: Enzyme‐inhibitor (EI), enzyme complex with a regulatory or accessory chain (ER), enzyme‐substrate (ES), others, receptor‐containing (OR); others, miscellaneous (OX)
