Figure 1: Slide-seq multiplex imaging workflow.
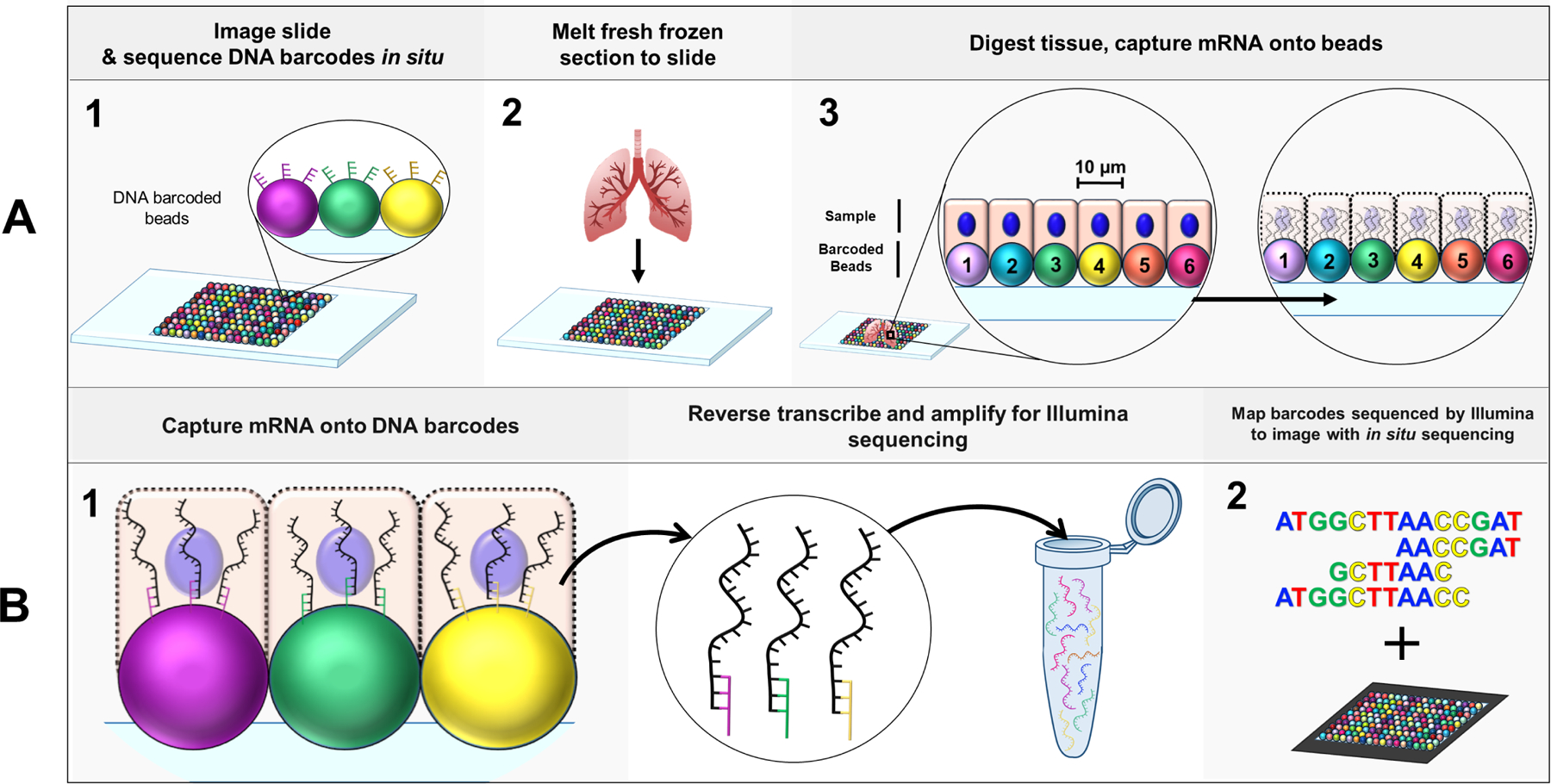
(A) Schematic of initial Slide-seq steps (A1) A rubber coated coverslip is covered in a monolayer of 10µm micro-particles (“beads”) to create a “puck”. Each bead is conjugated to its own unique barcode whose sequence is determined in situ via SOLiD (Sequencing by Oligonucleotide Ligation and Detection) for high resolution localization of each bead on the puck. (A2) Fresh frozen tissue can then be melted onto the puck, (A3) where the cells (approximately one cell/bead) overlap with the DNA barcoded beads. The tissue is digested and the mRNA of each cell is captured onto the DNA barcodes residing directly beneath the respective cell. (B) Schematic of library preparation for Slide-seq. (B1) mRNA are reverse transcribed to incorporate in a 3’-end, barcoded RNA-seq library preparation. Products are then amplified and undergo Illumina sequencing. (B2) Transcript profiles associated with a barcode sequence obtained from Illumina sequencing are mapped to their specific tissue location by matching the Illumina data to the barcode sequence from the initial in situ sequencing step in A1 [52].
